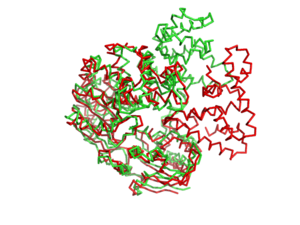
PcrA helicase
From Proteopedia
(→The Superfamily 1 (SF1)) |
|||
| (60 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image: | + | [[Image:1jprto3jpr.png|left|300px]] |
<!-- | <!-- | ||
| Line 13: | Line 13: | ||
''' | ''' | ||
| - | == What is a Helicase? == | + | == What is a Helicase? == |
| - | + | ||
| + | {{STRUCTURE_1pjr| PDB=1pjr | SIZE=400| SCENE= |right|CAPTION=PcrA , [[1pjr]] }} | ||
<scene name='User:Luis_E_Ramirez-Tapia/Sandbox_1/1pjrglobular/1'>Spacefill</scene> | <scene name='User:Luis_E_Ramirez-Tapia/Sandbox_1/1pjrglobular/1'>Spacefill</scene> | ||
Helicases are nucleic acid–dependent ATP-ases that are capable of unwinding DNA [http://en.wikipedia.org/wiki/DNA] or RNA [http://en.wikipedia.org/wiki/RNA] duplex substrates. As a consequence, they play roles in almost every process in cells that involves nucleic acids, including DNA replication and repair, transcription, translation, ribosome synthesis (1). | Helicases are nucleic acid–dependent ATP-ases that are capable of unwinding DNA [http://en.wikipedia.org/wiki/DNA] or RNA [http://en.wikipedia.org/wiki/RNA] duplex substrates. As a consequence, they play roles in almost every process in cells that involves nucleic acids, including DNA replication and repair, transcription, translation, ribosome synthesis (1). | ||
==PcrA a Simple Model for Helicases== | ==PcrA a Simple Model for Helicases== | ||
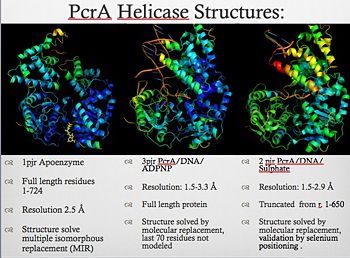
| - | PcrA is part of the replication machinery of the | + | [[Image:Pcr4_structures.jpg|thumb|350px|left|PcrA_Structure]] |
| - | + | PcrA is part of the replication machinery of the [http://en.wikipedia.org/wiki/Geobacillus_stearothermophilus Geobacillus stearothermophilus]a gram (+) bacteria, This helicase is part of the superfamily I of Helicases. Monomeric protein that is mainly <scene name='User:Luis_E_Ramirez-Tapia/Sandbox_1/Initial/1'>alfa helical</scene> has the <scene name='User:Luis_E_Ramirez-Tapia/Sandbox_2/1pjrconser/2'>highly conserved</scene> Rec domians. This helicase was reported as a mutation in the gen PcrA from [http://en.wikipedia.org/wiki/staphylococcu "Stapphylococcus aerous"], this mutation was related to a deficiency in the replication of a reporter plasmid.[http://www.ncbi.nlm.nih.gov/pubmed/8232203?ordinalpos=81&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum] | |
| - | + | <table align='right'><tr><td> </td><td>{{Template:ColorKey_ConSurf}}</td></tr></table> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{{clear}} | {{clear}} | ||
| - | == | + | ==PcrA Biochemistry== |
| - | + | PcrA is has an ATPas activityt which directionality is from 3' to 5' helicase strand separation reaction. The enzyme shows a specificity for the DNA substrate in gel mobility assays with the preferred substrate being one containing both single and double stranded regions of DNA. In contrast to Rep and UvrD from E. coli, there is not evidence for dimerisation of the enzyme using gel filtration, or by crosslinking in the presence of combinations of Mg2+, nucleotides and DNA. Moreover, kcat for ATP hydrolysis is constant over a large range of protein concentrations. Therefore, the protein appears to be monomeric under all conditions tested, including in the structure of two crystal forms of PcrA.[http://www.icnet.uk/labs/wigley/projects/helicase/35.html] | |
| + | [[Image:Wigleypcr4.jpg]] | ||
| - | + | ==PcrA Helicase Mechanism : The Mexican Wave== | |
| - | As can be seen when the chain is <scene name='Lac_repressor/1lbg_lac_repressor_with_dna/2'>colored with an N to C rainbow scheme</scene> | ||
| - | {{Template:ColorKey_N2CRainbow}} | ||
| - | each of the ligand-binding subdomains is made up of two discontinuous segments. | ||
| - | + | {{STRUCTURE_3pjr| PDB=3pjr | SIZE=400| SCENE= |right|CAPTION=PcrA complex with DNA and ATP, [[3pjr]] }} | |
| - | + | ||
| - | + | ||
| - | + | Professor Dale B. Wigley' group in 1996-1999 was able to crystalize the intermediate states from PcrA, giving solution to the controversy of what kind of mechanism this helicase has. [http://www.ncbi.nlm.nih.gov/pubmed/10199404ordinalpos=39&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum] | |
| - | + | Two crystal form of the enzyma, one couple with a 10 mer DNA and a non hydrolizable form of ATP (ATPnP) (pdb id: 3pjr [[3pjr]], <scene name='User:Luis_E_Ramirez-Tapia/Sandbox_2/3pjrinitial/1'>(Enzyme Subtrate Structure) </scene>and another a truncated form embebed in sulfate (pdb id: 2pjr [[2pjr]]<scene name='User:Luis_E_Ramirez-Tapia/Sandbox_2/2pjrinitial/1'>(Enzyme Product Structure)</scene>, give a light in a model for how ATP hydrolysis results in motor movement along ssDNA. In the figure below step 1 (top) is the ATP free (product) ssDNA conformation. The DNA bases are labelled arbitrarily. On binding ATP, F626 creates a new binding pocket for base 6. Likewise, F64 destroys an acceptor pocket for base 2, forcing it to move to the position occupied by base 1. After ATP hydrolysis, the grip on base 6 is released. When the Y257 pocket is re-opened due to movement of F64, bases 3-6 can now flip through the acceptor pockets to their new positions. This model predicts that each ATP hydrolysis event will advance PcrA one base along ssDNA.[http://www.icnet.uk/labs/wigley/projects/helicase/35.html] | |
| + | [[Image:Mexicanwave.jpg|thumb|170px|left|Inchworm or Mexicanwave model]] | ||
| + | [[Image:Snapshot_2008-12-03_14-11-31.jpg|thumb|400px|left|PcrA Movie]] | ||
| - | <--! scene with translucent :a is #11, but I didn't like it. --> | ||
| - | The C-terminal tetramerization helices tether two dimers, and thus the functional form of <scene name='Lac_repressor/1lbg_lac_repressor_with_dna/7'>lac repressor is a homo-tetramer</scene> with two {{Template:ColorKey_Composition_DNA}}-binding sites. | ||
| - | + | '''The link below show a movie with the principal characteristics of this protein as long with the inchworm model'''. [http://www.youtube.com/watch?v=fDwaWCkhgZI Pcr4 Helicase and Mexican Wave] | |
| + | {{Clear}} | ||
| - | == | + | ==The Superfamily 1 (SF1)== |
| - | + | PcrA share structural domains with the Rec helicases, like UvrD and RepD from E. coli, Superfamily 1 (SF1) helicases are probably the best characterized class, certainly from a structural perspective. All members characterized to date are bona fide helicases and α enzymes. Indeed, from their mode of translocation via the bases it is difficult to envisage how they could translocate along a duplex. However, they can have either A or B directional polarity. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | {{STRUCTURE_2is1| PDB=2is1 | SIZE=300| SCENE= |left|CAPTION=UVRD complex with DNA and sulfate, [[2is1]] }} | |
| + | {{STRUCTURE_1uaa| PDB=1uaa | SIZE=300| SCENE= |center|CAPTION=REP complex with DNA, [[1uaa]] }} | ||
| - | + | {{clear}} | |
| - | Upon recognizing the specific operator sequence, the non-specific binding converts to <scene name='Lac_repressor/1l1m_ca_specific_bindiing/3'>specific binding</scene> (derived<ref name='alphac' /> from [[1l1m]], 20 [[NMR Ensembles of Models|NMR models]]). During this conversion, the hinge region changes from disordered loops to {{Template:ColorKey_Helix}} (<scene name='Lac_repressor/1l1m_ca_specific_bindiing/4'>highlight new helices</scene>), which bind in the minor groove of the DNA. This binding opens the minor groove, bending the <font color='#ae00ff'><b>DNA double helix</b></font>. <scene name='Lac_repressor/1l1m_ca_specific_bindiing/6'>Animating</scene> these can be compared with the animation of the non-specific binding. {{Template:Button Toggle Animation}} | ||
| - | ====Morph of Conversion==== | ||
| - | The <scene name='Morphs/1osl_19_1l1m_9_morph/4'>changes during conversion from non-specific to specific binding</scene> can be seen more easily when they are animated smoothly by [[Morphs|morphing]]. (The methods used to create this morph are given in [[Lac repressor morph methods]].) Note the bending of the DNA, with the widening of the central minor groove on the convex aspect. Also note the conversion of the disulfide-bonded hinge region loops to alpha helices. (The displayed secondary structure is calculated for each model in the morph interpolation.) | ||
| - | The specific recognition of the lac operator sequence in the DNA occurs largely though [[Hydrogen bonds|hydrogen bonds]]. <scene name='Lac_repressor/1osl_14_1l1m_9_morph_hbonds/1'>Formation of hydrogen bonds that recognize the operator sequence</scene> is illustrated in this rendering of the morph. Shown are hydrogen bonds involving Arg22.N-eta2 and Tyr18.OH interacting with DNA base oxygens in the major groove, and Ala53.O interacting with a DNA base nitrogen in the minor groove. (Not all of the relevant hydrogen bonds are shown; see [[Lac repressor morph methods|Methods]].) {{Template:Button Toggle Animation}} | ||
| - | Test: <scene name='Lac_repressor/1osl_19_1l1m_9_morph/1'>bad morph scene</scene> | ||
| - | == | + | ==About this Structure== |
| - | + | 1PJR is a [[Single protein]] structure of sequence from [http://en.wikipedia.org/wiki/Geobacillus_stearothermophilus Geobacillus stearothermophilus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1PJR OCA]. | |
| - | [ | + | |
| - | + | ==3D structures of helicase== | |
| - | + | ||
| - | + | [[Helicase]] | |
| - | + | ==Additional Resources== | |
| - | + | For additional information, see: [[DNA Replication, Repair, and Recombination]] | |
| - | + | <br /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Reference== | ==Reference== | ||
| Line 128: | Line 82: | ||
''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Jul 28 01:38:49 2008'' | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Jul 28 01:38:49 2008'' | ||
| + | |||
| + | ^ Johnson DS, Bai L, Smith BY, Patel SS, Wang MD (2007). "Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped t7 helicase". Cell 129 (7): 1299–309. doi:10.1016/j.cell.2007.04.038. PMID 17604719. | ||
| + | ^ a b "Researchers solve mystery of how DNA strands separate" (2007-07-03). Retrieved on 2007-07-05. | ||
| + | ^ Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco Jr I, Pylr AM, Bustamante C, "RNA Translocation and Unwinding Mechanism of HCV NS3 Helicase and its Coordination by ATP", Nature. 2006 Jan 5; 439: 105-108. | ||
| + | Anand SP, Zheng H, Bianco PR, Leuba SH, Khan SA. DNA helicase activity of PcrA is not required for displacement of RecA protein from DNA or inhibition of RecA-mediated DNA strand exchange. Journal of Bacteriology (2007) 189 (12):4502-4509. | ||
| + | Bird L, Subramanya HS, Wigley DB, "Helicases: a unifying structural theme?", Current Opinion in Structural Biology. 1998 Feb; 8 (1): 14-18. | ||
| + | Betterton MD, Julicher F, "Opening of nucleic-acid double strands by helicases: active versus passive opening.", Physical Review E. 2005 Jan; 71 (1): 011904. | ||
Current revision
Contents |
What is a Helicase?
| |||||||||
| PcrA , 1pjr | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene: | PCRA (Geobacillus stearothermophilus) | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Helicases are nucleic acid–dependent ATP-ases that are capable of unwinding DNA [1] or RNA [2] duplex substrates. As a consequence, they play roles in almost every process in cells that involves nucleic acids, including DNA replication and repair, transcription, translation, ribosome synthesis (1).
PcrA a Simple Model for Helicases
PcrA is part of the replication machinery of the Geobacillus stearothermophilusa gram (+) bacteria, This helicase is part of the superfamily I of Helicases. Monomeric protein that is mainly has the Rec domians. This helicase was reported as a mutation in the gen PcrA from "Stapphylococcus aerous", this mutation was related to a deficiency in the replication of a reporter plasmid.[3]
 |
PcrA Biochemistry
PcrA is has an ATPas activityt which directionality is from 3' to 5' helicase strand separation reaction. The enzyme shows a specificity for the DNA substrate in gel mobility assays with the preferred substrate being one containing both single and double stranded regions of DNA. In contrast to Rep and UvrD from E. coli, there is not evidence for dimerisation of the enzyme using gel filtration, or by crosslinking in the presence of combinations of Mg2+, nucleotides and DNA. Moreover, kcat for ATP hydrolysis is constant over a large range of protein concentrations. Therefore, the protein appears to be monomeric under all conditions tested, including in the structure of two crystal forms of PcrA.[4]
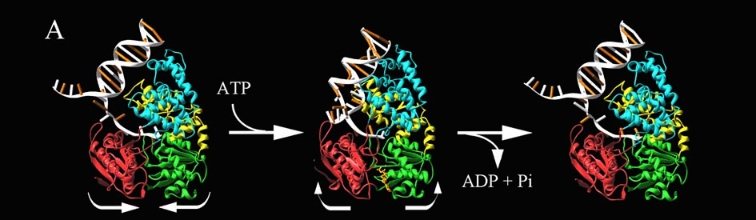
PcrA Helicase Mechanism : The Mexican Wave
| |||||||||
| PcrA complex with DNA and ATP, 3pjr | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Professor Dale B. Wigley' group in 1996-1999 was able to crystalize the intermediate states from PcrA, giving solution to the controversy of what kind of mechanism this helicase has. [5]
Two crystal form of the enzyma, one couple with a 10 mer DNA and a non hydrolizable form of ATP (ATPnP) (pdb id: 3pjr 3pjr, and another a truncated form embebed in sulfate (pdb id: 2pjr 2pjr, give a light in a model for how ATP hydrolysis results in motor movement along ssDNA. In the figure below step 1 (top) is the ATP free (product) ssDNA conformation. The DNA bases are labelled arbitrarily. On binding ATP, F626 creates a new binding pocket for base 6. Likewise, F64 destroys an acceptor pocket for base 2, forcing it to move to the position occupied by base 1. After ATP hydrolysis, the grip on base 6 is released. When the Y257 pocket is re-opened due to movement of F64, bases 3-6 can now flip through the acceptor pockets to their new positions. This model predicts that each ATP hydrolysis event will advance PcrA one base along ssDNA.[6]
The link below show a movie with the principal characteristics of this protein as long with the inchworm model. Pcr4 Helicase and Mexican Wave
The Superfamily 1 (SF1)
PcrA share structural domains with the Rec helicases, like UvrD and RepD from E. coli, Superfamily 1 (SF1) helicases are probably the best characterized class, certainly from a structural perspective. All members characterized to date are bona fide helicases and α enzymes. Indeed, from their mode of translocation via the bases it is difficult to envisage how they could translocate along a duplex. However, they can have either A or B directional polarity.
| |||||||||
| UVRD complex with DNA and sulfate, 2is1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | uvrD (Escherichia coli) | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
| |||||||||
| REP complex with DNA, 1uaa | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
About this Structure
1PJR is a Single protein structure of sequence from Geobacillus stearothermophilus. Full crystallographic information is available from OCA.
3D structures of helicase
Additional Resources
For additional information, see: DNA Replication, Repair, and Recombination
Reference
Crystal structure of a DExx box DNA helicase., Subramanya HS, Bird LE, Brannigan JA, Wigley DB, Nature. 1996 Nov 28;384(6607):379-83. PMID:8934527
Page seeded by OCA on Mon Jul 28 01:38:49 2008
^ Johnson DS, Bai L, Smith BY, Patel SS, Wang MD (2007). "Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped t7 helicase". Cell 129 (7): 1299–309. doi:10.1016/j.cell.2007.04.038. PMID 17604719. ^ a b "Researchers solve mystery of how DNA strands separate" (2007-07-03). Retrieved on 2007-07-05. ^ Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco Jr I, Pylr AM, Bustamante C, "RNA Translocation and Unwinding Mechanism of HCV NS3 Helicase and its Coordination by ATP", Nature. 2006 Jan 5; 439: 105-108. Anand SP, Zheng H, Bianco PR, Leuba SH, Khan SA. DNA helicase activity of PcrA is not required for displacement of RecA protein from DNA or inhibition of RecA-mediated DNA strand exchange. Journal of Bacteriology (2007) 189 (12):4502-4509. Bird L, Subramanya HS, Wigley DB, "Helicases: a unifying structural theme?", Current Opinion in Structural Biology. 1998 Feb; 8 (1): 14-18. Betterton MD, Julicher F, "Opening of nucleic-acid double strands by helicases: active versus passive opening.", Physical Review E. 2005 Jan; 71 (1): 011904.
Proteopedia Page Contributors and Editors (what is this?)
Luis E Ramirez-Tapia, Michal Harel, Eran Hodis, David Canner