User:Jeremy C. Caylor/Sandbox 1
From Proteopedia
| (2 intermediate revisions not shown.) | |||
| Line 16: | Line 16: | ||
1. Hydrogen bonding with RNA bases: | 1. Hydrogen bonding with RNA bases: | ||
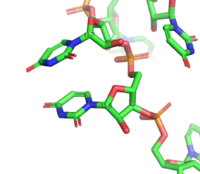
| - | There are numerous hydrogen bonds between the residues and the nucleotide bases. RRM2 creates three hydrogen bonds with U3 and G4 with residues <scene name='78/782600/H-bond_u3_bases/3'>Asn217 and Arg252</scene> and residue <scene name='78/782600/Ala289_g4/1'>Ala289</scene>. It also forms two hydrogen bonds with U5 with residues <scene name='78/782600/Rrm2_res_hbond_w_bases/ | + | There are numerous hydrogen bonds between the residues and the nucleotide bases. RRM2 creates three hydrogen bonds with U3 and G4 with residues <scene name='78/782600/H-bond_u3_bases/3'>Asn217 and Arg252</scene> and residue <scene name='78/782600/Ala289_g4/1'>Ala289</scene>. It also forms two hydrogen bonds with U5 with residues <scene name='78/782600/Rrm2_res_hbond_w_bases/5'>Aln239 and Asn212</scene>. RRM1 creates five hydrogen bonds with U6, U7, and U8 with residues <scene name='78/782600/Rrm1_res_h-bond_w_bases/6'>Arg195, Gln134, and Ser165</scene>. It also creates seven hydrogens bonds to U9, U10, and U11 with residues <scene name='78/782600/U9_u10/1'>Gly204, Arg202, Lys197</scene> and residues <scene name='78/782600/U11/1'>Arg155 and Asn126</scene><ref name="Handa" />. |
2. Hydrogen bonding with the RNA backbone: | 2. Hydrogen bonding with the RNA backbone: | ||
| Line 30: | Line 30: | ||
==Autoregulation== | ==Autoregulation== | ||
| - | [[Image: | + | [[Image:Screen Shot 2018-04-26 at 4.55.49 PM.png|300 px|right|thumb|Figure 4. Alternative splicing mechanism for autoregulation of Sxl. The presence of Sxl causes the alternative splicing of exon 3 in the female mRNA transcript. The retention of exon 3, along with the premature stop codon it contains, leads to a truncated and inactive Sxl protein in males.]] |
Sxl controls its own levels of expression via positive and negative [https://en.wikipedia.org/wiki/Autoregulation autoregulation]. Sxl binds its own pre-mRNA transcript in a similar manner as its downstream targets, Tra and Msl2. Through binding to its recognition element, Sxl causes a 3’ splice site to be skipped. [https://en.wikipedia.org/wiki/Alternative_splicing Alternative splicing] occurs utilizing a 3’ splice site further downstream, cleaving out a premature stop codon within Exon 3 and preventing truncation and inactivation<ref name="Black" /> (Figure 4). This is a pathway of positive autoregulation, as functional Sxl protein must be present to cause proper processing of the pre-mRNA. The negative autoregulation pathway of Sxl proceeds via repression of its own translation. The Sxl transcript contains the target polyuridine sequence within its 3’UTR. Sxl binds this target, and blocks translation<ref name="Black" />. Negative autoregulation allows maintenance of a stable and standard Sxl protein concentration. An excess of Sxl increases the degree of translation repression because more Sxl protein are present to potentially bind at the 3’UTR, while a shortage allows for more unrepressed translation. | Sxl controls its own levels of expression via positive and negative [https://en.wikipedia.org/wiki/Autoregulation autoregulation]. Sxl binds its own pre-mRNA transcript in a similar manner as its downstream targets, Tra and Msl2. Through binding to its recognition element, Sxl causes a 3’ splice site to be skipped. [https://en.wikipedia.org/wiki/Alternative_splicing Alternative splicing] occurs utilizing a 3’ splice site further downstream, cleaving out a premature stop codon within Exon 3 and preventing truncation and inactivation<ref name="Black" /> (Figure 4). This is a pathway of positive autoregulation, as functional Sxl protein must be present to cause proper processing of the pre-mRNA. The negative autoregulation pathway of Sxl proceeds via repression of its own translation. The Sxl transcript contains the target polyuridine sequence within its 3’UTR. Sxl binds this target, and blocks translation<ref name="Black" />. Negative autoregulation allows maintenance of a stable and standard Sxl protein concentration. An excess of Sxl increases the degree of translation repression because more Sxl protein are present to potentially bind at the 3’UTR, while a shortage allows for more unrepressed translation. | ||
Current revision
Contents |
Sex lethal: RNA-binding protein
Introduction
Structure
| |||||||||||
Autoregulation
Sxl controls its own levels of expression via positive and negative autoregulation. Sxl binds its own pre-mRNA transcript in a similar manner as its downstream targets, Tra and Msl2. Through binding to its recognition element, Sxl causes a 3’ splice site to be skipped. Alternative splicing occurs utilizing a 3’ splice site further downstream, cleaving out a premature stop codon within Exon 3 and preventing truncation and inactivation[1] (Figure 4). This is a pathway of positive autoregulation, as functional Sxl protein must be present to cause proper processing of the pre-mRNA. The negative autoregulation pathway of Sxl proceeds via repression of its own translation. The Sxl transcript contains the target polyuridine sequence within its 3’UTR. Sxl binds this target, and blocks translation[1]. Negative autoregulation allows maintenance of a stable and standard Sxl protein concentration. An excess of Sxl increases the degree of translation repression because more Sxl protein are present to potentially bind at the 3’UTR, while a shortage allows for more unrepressed translation.
Mutation
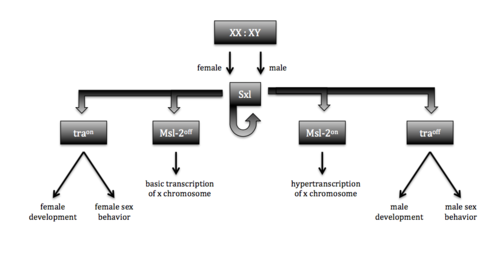
In fruit flies, dosage compensation is achieved by hyperexpression of the single X chromosome found in the male genome. The single X chromosome is hyperexpressed and regulated by a group of genes known as the Male-specific lethal genes. The genes involved work together to produce a complex known as the Male-specific lethal complex (Msl complex). One gene that is important in forming this complex is the gene that codes for the Msl-2 protein. Without this protein, the Msl complex cannot form [5]. Males exclusively make the Msl-2 protein and therefore exclusively create the Msl complex. Females cannot make the complex because the Sxl protein prevents the production of the Msl-2 protein. It does this by alternatively splicing an intron near the 5’ UTR. In males this intron is included and in females it is not [5]. Mutations of the Sxl protein can affect the formation of this complex and lead to death for females—hence the name sex lethal. If the Sxl gene is mutated in females, the formation of the Msl-2 complex will not be inhibited. If there is a functional Msl-2 complex in females then both of the X chromosomes will be hyperexpressed [5]. The overexpression of genes on the X chromosome is a fatal phenomenon ultimately caused by a mutation of the Sxl protein. Although a mutation to the Sxl gene is harmful to females, it has no effect on males because the functional Sxl gene is not expressed—a mutation in males has no phenotypic effect.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291-336. doi: 10.1146/annurev.biochem.72.121801.161720., Epub 2003 Feb 27. PMID:12626338 doi:http://dx.doi.org/10.1146/annurev.biochem.72.121801.161720
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999 Apr 15;398(6728):579-85. PMID:10217141 doi:10.1038/19242
- ↑ Georgiev P, Chlamydas S, Akhtar A. Drosophila dosage compensation. Fly 2011;5(2):147-154. https://doi.org/10.4161/fly.5.2.14934
- ↑ 4.0 4.1 Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008 Jun;18(3):290-8. doi: 10.1016/j.sbi.2008.04.002. PMID:18515081 doi:http://dx.doi.org/10.1016/j.sbi.2008.04.002
- ↑ 5.0 5.1 5.2 Penalva L, Sanchez L. RNA Binding Protein Sex-Lethal (Sxl) and Control of Drosophila Sex Determination and Dosage Compensation. Microbiol Mol Biol Rev.;67(3):343-356. doi: 10.1128/MMBR.67.3.343–359.2003