User:Scott H. Vanson/Sandbox 1
From Proteopedia
| Line 4: | Line 4: | ||
== General Description == | == General Description == | ||
| - | Human polymerase θ (pol θ) is large, 290kD enzyme consisting of three distinct domains <ref>PMID:23219161</ref><ref name="Seki">PMID:14576298</ref>. An N-terminal helicase-like domain, whose exact cellular functions are a topic of on-going debate and research<ref>PMID:29058711</ref><ref>PMID:29444826</ref>, is linked to a C-terminal, family A DNA polymerase domain by a large and disordered central region<ref name="Seki" />. Notably, pol θ is the only known human polymerase to contain a polymerase and helicase domain in one molecule<ref name="Zahn">PMID: | + | Human polymerase θ (pol θ) is large, 290kD enzyme consisting of three distinct domains <ref>PMID:23219161</ref><ref name="Seki">PMID:14576298</ref>. An N-terminal helicase-like domain, whose exact cellular functions are a topic of on-going debate and research<ref>PMID:29058711</ref><ref>PMID:29444826</ref>, is linked to a C-terminal, family A DNA polymerase domain by a large and disordered central region<ref name="Seki" />. Notably, pol θ is the only known human polymerase to contain a polymerase and helicase domain in one molecule<ref name="Zahn">PMID:25775267</ref>. Crystal structures have been solved for the apo form of the [http://www.rcsb.org/structure/5a9j helicase-like domain] and the ternary complex of the [http://http://www.rcsb.org/structure/4X0P polymerase domain]. The focus of this wiki is the polymerase domain. |
| + | [[Image:2D_PolQ_scheme.png|thumb|left|upright=4]] | ||
| - | + | ===Cellular Functions=== | |
| - | + | Pol θ is thought to promote overall genomic stability by performing several distinct cellular functions. The primary role of the enzyme is to repair of double-stranded DNA breaks as the key enzyme in an error-prone, non-homologous end-joining pathway called alternative end-joining<ref name="Instability">PubMed:25275444</ref> or theta-mediated end-joining. Other functions include translesion synthesis, the ability of the polymerase to bypass and extend past a site of oxidative DNA damage<ref>PubMed:24648516</ref>, base excision repair <ref>PubMed:21917855</ref>, and possibly DNA replication timing <ref>PubMed:24989122</ref>. Pol θ has the specialized ability to extend DNA from minimally-paired primers (termed microhomologous){{cn}}. Repair by this enzyme is considered to error-prone due to its tendency to add or delete short indels <ref name="Instability" />. | |
| + | ===Disease=== | ||
| + | Several types of cancer, such as breast, ovarian, and oral carcinomas, have shown significantly higher expression levels of pol θ and correlate to poorer patient outcomes<ref name="Ceccaldi">PMID:25642963</ref><ref>PubMed:20624954</ref><ref>PubMed:22987617</ref>. Genomic studies have shown that more than half of epithelial ovarian cancers have defects in the error-free repair pathway of homologous recombination<ref>PMID:21720365</ref> and, as a result, have an increased dependence on theta-mediated end-joining <ref name="Ceccaldi" />. Double-stranded break repair by pol θ may be thought of as a "backup" pathway which cells depend on more when the machinery involved in homologous recombination is compromised or otherwise unavailable. This enzyme has been identified as a potential therapeutic target due to overexpression in cancers in combination with studies that have shown inhibition of pol θ to sensitize human and mouse cells to radiation and chemical agents which induce double-stranded breaks<ref name="Ceccaldi" /><ref>PubMed:19630521</ref><ref>PubMed:20233878</ref>. | ||
== Structural Highlights == | == Structural Highlights == | ||
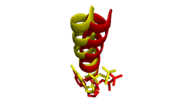
| - | Two crystal structures of the polymerase domain have been solved bound | + | Two crystal structures of the polymerase domain have been solved bound to DNA to a resolution of 3.9Å; inserting ddATP opposite tetrahydrofuran (THF, representing an abasic site) and inserting ddGTP opposite dCMP<ref name="Zahn" />. An overall assessment of the structures display the canonical <scene name='78/786633/Subdomains/2'>exonuclease, thumb, palm, and fingers subdomains</scene>. The DNA is thought to "sit" on the palm and is enclosed by the thumb and fingers via the minor groove. The palm houses the active site and the fingers move from an open, unbound position to a closed position when bound to DNA, promoting synthesis. The closed conformation of the polymerase domain becomes obvious when compared to the <scene name='78/786633/Taq_pol_i_open/1'>open conformation of a homologous structure</scene>, such as [http://www.rcsb.org/structure/1KTQ Thermus aquaticus DNA polymerase I]. |
===ddATP Opposite THF=== | ===ddATP Opposite THF=== | ||
| Line 29: | Line 32: | ||
===Translesion Synthesis=== | ===Translesion Synthesis=== | ||
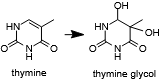
| - | An R2254V variant was made to investigate the importance of this residue in pol θ's ability to extend single-stranded DNA and bypass abasic sites and bulky thymine glycol lesions.[[Image:Thymine_glycol.png|thumb|left|Thymine may oxidized to form a bulky lesion that must be repaired or bypassed.]] Bacterial family A polymerases which do not have this ability retain a | + | An R2254V variant was made to investigate the importance of this residue in pol θ's ability to extend single-stranded DNA and bypass abasic sites and bulky thymine glycol lesions.[[Image:Thymine_glycol.png|thumb|left|Thymine may oxidized to form a bulky lesion that must be repaired or bypassed.]] Bacterial family A polymerases which do not have this ability retain a valine or leucine at the equivalent position. The R2254V mutant retained its ability to extend double-stranded but not single-stranded DNA and also was not able to bypass abasic sites or thymine glycol. These findings indicated to the authors that <scene name='78/786633/R2254/1'>the salt bridge between R2254 and the 3'-terminal phosphate of the primer</scene> is required to compensate for the missing contacts to the template strand due to a lesion<ref name="Zahn" />. |
====Sticky Thumb==== | ====Sticky Thumb==== | ||
In addition to R2254, additional upstream contacts to the primer DNA strand mediated by <scene name='78/786633/Lys_2181/1'>K2181</scene> and <scene name='78/786633/Arg2202/1'>R2202</scene> of the thumb subdomain can be observed. These three salt bridges provide specialized contacts that are not observed in other family A polymerases, in addition to R2201 and R2315 which are also present in [http://www.rcsb.org/structure/4XVK pol ν] and pol I. Alanine substitutions of K2181, R2202, and R2254 resulted in inhibition of pol θ's ability to bypass an abasic site or thymine glycol. These findings prompted the authors to assert that this <scene name='78/786633/Sticky_thumb/1'>specialized thumb domain</scene> may be responsible for the heightened ability of pol θ to synthesize across lesions and to extend from minimally-paired primers<ref name="Zahn" />. | In addition to R2254, additional upstream contacts to the primer DNA strand mediated by <scene name='78/786633/Lys_2181/1'>K2181</scene> and <scene name='78/786633/Arg2202/1'>R2202</scene> of the thumb subdomain can be observed. These three salt bridges provide specialized contacts that are not observed in other family A polymerases, in addition to R2201 and R2315 which are also present in [http://www.rcsb.org/structure/4XVK pol ν] and pol I. Alanine substitutions of K2181, R2202, and R2254 resulted in inhibition of pol θ's ability to bypass an abasic site or thymine glycol. These findings prompted the authors to assert that this <scene name='78/786633/Sticky_thumb/1'>specialized thumb domain</scene> may be responsible for the heightened ability of pol θ to synthesize across lesions and to extend from minimally-paired primers<ref name="Zahn" />. | ||
| - | |||
| - | == Related Proteins == | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| - | 1. Yousefzadeh MJ, Wood RD. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair (Amst). 2013; 12:1–9. [PubMed: 23219161] | ||
| - | 2. Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003; 31:6117–6126. [PubMed: 14576298] | ||
| - | 3. Sfeir RPA | ||
| - | 4. Pomerantz Helicase PMID:29444826 | ||
| - | 5. Zahn | ||
6. Newman | 6. Newman | ||
6. Yousefzadeh MJ, et al. Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ. PLoS Genet. 2014; 10:e1004654. [PubMed: 25275444] | 6. Yousefzadeh MJ, et al. Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ. PLoS Genet. 2014; 10:e1004654. [PubMed: 25275444] | ||
Revision as of 20:15, 1 May 2018
DNA Polymerase θ
| |||||||||||
References
6. Newman 6. Yousefzadeh MJ, et al. Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ. PLoS Genet. 2014; 10:e1004654. [PubMed: 25275444] 7. Yoon JH, Roy Choudhury J, Park J, Prakash S, Prakash L. A role for DNA polymerase theta in promoting replication through oxidative DNA lesion, thymine glycol, in human cells. J Biol Chem. 2014; 289:13177–13185. [PubMed: 24648516] 8. Asagoshi K, et al. Single-nucleotide base excision repair DNA polymerase activity in C. elegans in the absence of DNA polymerase beta. Nucleic Acids Res. 2012; 40:670–681. [PubMed: 21917855] 9. Fernandez-Vidal A, et al. A role for DNA polymerase theta in the timing of DNA replication. Nat Commun. 2014; 5:4285. [PubMed: 24989122] 10. Ceccaldi 11. Lemee F, et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc Natl Acad Sci U S A. 2010; 107:13390–13395. [PubMed: 20624954] 12. Lessa RC, et al. Identification of upregulated genes in oral squamous cell carcinomas. Head Neck. 2013; 35:1475–1481. [PubMed: 22987617] 13. Cancer Genome Atlas Research, N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011; 474:609–615.10.1038/nature10166 14. Goff JP, et al. Lack of DNA polymerase theta (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat Res. 2009; 172:165–174. [PubMed: 19630521] 15. Higgins GS, et al. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 2010; 70:2984–2993. [PubMed: 20233878]
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Yousefzadeh MJ, Wood RD. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair (Amst). 2013 Jan 1;12(1):1-9. doi: 10.1016/j.dnarep.2012.10.004. Epub , 2012 Dec 4. PMID:23219161 doi:http://dx.doi.org/10.1016/j.dnarep.2012.10.004
- ↑ 4.0 4.1 Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003 Nov 1;31(21):6117-26. PMID:14576298
- ↑ Mateos-Gomez PA, Kent T, Deng SK, McDevitt S, Kashkina E, Hoang TM, Pomerantz RT, Sfeir A. The helicase domain of Poltheta counteracts RPA to promote alt-NHEJ. Nat Struct Mol Biol. 2017 Dec;24(12):1116-1123. doi: 10.1038/nsmb.3494. Epub 2017, Oct 23. PMID:29058711 doi:http://dx.doi.org/10.1038/nsmb.3494
- ↑ Ozdemir AY, Rusanov T, Kent T, Siddique LA, Pomerantz RT. Polymerase theta-helicase efficiently unwinds DNA and RNA-DNA hybrids. J Biol Chem. 2018 Apr 6;293(14):5259-5269. doi: 10.1074/jbc.RA117.000565. Epub, 2018 Feb 14. PMID:29444826 doi:http://dx.doi.org/10.1074/jbc.RA117.000565
- ↑ 7.0 7.1 7.2 7.3 7.4 Zahn KE, Averill AM, Aller P, Wood RD, Doublie S. Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat Struct Mol Biol. 2015 Mar 16. doi: 10.1038/nsmb.2993. PMID:25775267 doi:http://dx.doi.org/10.1038/nsmb.2993
- ↑ 8.0 8.1 PubMed:25275444
- ↑ PubMed:24648516
- ↑ PubMed:21917855
- ↑ PubMed:24989122
- ↑ 12.0 12.1 12.2 Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, O'Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, Yusufzai T, D'Andrea AD. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015 Feb 12;518(7538):258-62. doi: 10.1038/nature14184. Epub 2015 Feb 2. PMID:25642963 doi:http://dx.doi.org/10.1038/nature14184
- ↑ PubMed:20624954
- ↑ PubMed:22987617
- ↑ . Integrated genomic analyses of ovarian carcinoma. Nature. 2011 Jun 29;474(7353):609-15. doi: 10.1038/nature10166. PMID:21720365 doi:http://dx.doi.org/10.1038/nature10166
- ↑ PubMed:19630521
- ↑ PubMed:20233878