Xylosidase
From Proteopedia
(Difference between revisions)
| (14 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
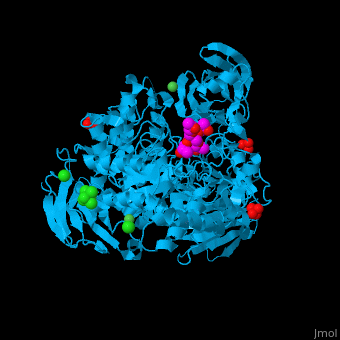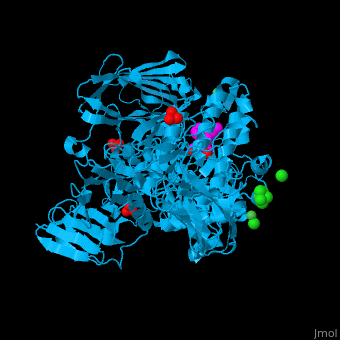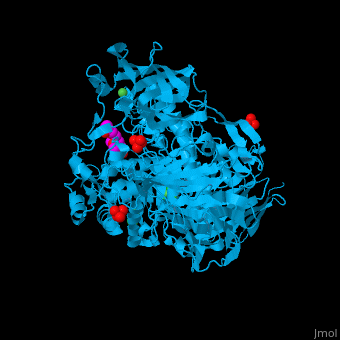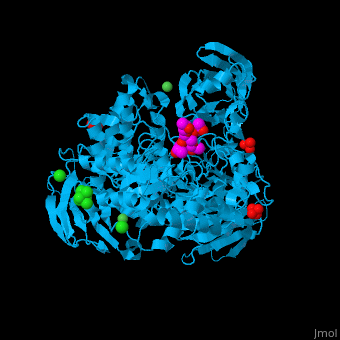
| - | <StructureSection load=' | + | <StructureSection load='' size='350' side='right' caption='Structure of α-xylosidase family 31 complex with pentaerythritol propoxylate, sulfate, Ni+2 (small green) and Cl- (large green) ions (PDB entry [[2xvl]])' scene='55/551220/Cv/1'> |
| + | __TOC__ | ||
== Function == | == Function == | ||
| - | '''Xylosidase''' (XYL) catalyzes the hydrolysis of α- or β-glycosidic linkages. '''β-XYL''' catalyzes the endo-hydrolysis of 1,4-β-D-glycosidic or endo-hydrolysis of 1,3-β-D-glycosidic linkage<ref>PMID:15283123</ref>. '''α-XYL''' catalyzes the | + | '''Xylosidase''' (XYL) catalyzes the hydrolysis of α- or β-glycosidic linkages. |
| + | |||
| + | *'''α-XYL''' catalyzes the exo-hydrolysis of 1,4-β-D-glycosidic or exo-hydrolysis of 1,3-β-D-glycosidic linkage from the non-reducing termini of xylans<ref>PMID:10801892</ref>. XYL catalyzes the conversion of cellulose and hemicellulose xylan to xylose. | ||
| + | *'''β-XYL''' catalyzes the endo-hydrolysis of 1,4-β-D-glycosidic or endo-hydrolysis of 1,3-β-D-glycosidic linkage<ref>PMID:15283123</ref>. | ||
| + | *'''α-XYL family 31''' catalyzes the hydrolysis of α-xylosyl from isoprimeverose<ref>PMID:35441671</ref>. | ||
| + | *'''β-XYL family 3''' catalyzes the endo-hydrolysis of 1,4-β-D-glycosidic or endo-hydrolysis of 1,3-β-D-glycosidic linkage<ref>PMID:35120929</ref>. | ||
| + | *'''β-XYL family 43''' and '''β-XYL family 52''' degrade xylooligosaccharides<ref>PMID:26494804</ref>, <ref>PMID:12738774</ref>. | ||
| + | *'''β-XYL family 120''' degrades xylobiose and xylotriose<ref>PMID:31698702</ref>. | ||
| + | *'''β-XYL/glucosidase''' bifunctional enzyme active on beta-xylosides and beta-glucosides. | ||
== Structural highlights == | == Structural highlights == | ||
| - | The active site of α-xylosidase contains the xyloglucan and nucleophilic water<ref>PMID:21426303</ref>. | + | The <scene name='55/551220/Cv/5'>active site of α-xylosidase contains the xyloglucan and nucleophilic water</scene><ref>PMID:21426303</ref>. Water molecules are shown as red spheres. <scene name='55/551220/Cv/6'>Ni/Cl complex</scene>. |
| - | </ | + | |
| - | + | ||
== 3D Structures of xylosidase == | == 3D Structures of xylosidase == | ||
| + | [[Xylosidase 3D structures]] | ||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | |||
| - | *α-xylosidase | ||
| - | |||
| - | **[[5jou]] – BoXYL – ''Bacterioides ovatus'' <BR /> | ||
| - | **[[5jov]] – BoXYL + piperazine derivative <BR /> | ||
| - | |||
| - | *α-xylosidase family 31 | ||
| - | |||
| - | **[[2xvg]], [[2xvk]] – CjXYL – ''Cellvibrio japonicus'' <BR /> | ||
| - | **[[2xvl]] – CjXYL + xylobiose <BR /> | ||
| - | **[[1xsi]], [[1xsj]], [[1we5]] – EcXYL – ''Escherichia coli''<br /> | ||
| - | **[[1xsk]] – EcXYL + intermediate<br /> | ||
| - | |||
| - | *β-xylosidase | ||
| - | |||
| - | **[[1yrz]] – XYL – ''Bacillus halodurans'' <BR /> | ||
| - | **[[1yif]] – XYL – ''Bacillus subtilis'' <BR /> | ||
| - | **[[4mlg]] – XYL – uncultured <BR /> | ||
| - | **[[3c2u]] – XYL + propane derivative – ''Selenomonas ruminantium'' <BR /> | ||
| - | |||
| - | *β-xylosidase family 3 | ||
| - | |||
| - | **[[5a7m]] – HjXYL – ''Hypocrea jecorina'' <BR /> | ||
| - | **[[5ae6]] – HjXYL + thioxylobiose <BR /> | ||
| - | |||
| - | *β-xylosidase family 39 | ||
| - | |||
| - | **[[1px8]], [[1uhv]] – TsXYL – ''Thermoanaerobacterium saccharolyticum'' <BR /> | ||
| - | **[[4ekj]] – XYL – ''Caulobacter vibrioides''<BR /> | ||
| - | **[[4m29]] – XYL – ''Caulobacter crscentus''<BR /> | ||
| - | **[[2bs9]], [[1w91]], [[4rhh]] – BsXYL – ''Bacillus stearothermophilus'' <BR /> | ||
| - | **[[2bfg]] – BsXYL + xylobiose <BR /> | ||
| - | |||
| - | *β-xylosidase family 43 | ||
| - | |||
| - | **[[1y7b]], [[1yi7]], [[3k1u]] – CaXYL – ''Clostridium acetobutylicum'' <BR /> | ||
| - | **[[3qed]], [[3qee]] – CjXYL <BR /> | ||
| - | **[[2exh]] – BsXYL <BR /> | ||
| - | **[[2exi]] – BsXYL (mutant) <BR /> | ||
| - | **[[2exj]] – BsXYL + xylobiose <BR /> | ||
| - | **[[2exk]] – BsXYL (mutant) + xylobiose <BR /> | ||
| - | **[[3qef]] – CaXYL (mutant) + arabinofuranose <BR /> | ||
| - | |||
| - | *β-xylosidase family 52 | ||
| - | |||
| - | **[[4c1o]] – BtXYL – ''Bacillus thermoglucosidasius'' <BR /> | ||
| - | **[[4c1p]] – BtXYL + xylobiose <BR /> | ||
| - | |||
| - | *β-xylosidase family 120 | ||
| - | |||
| - | **[[3vst]] – TsXYL <BR /> | ||
| - | **[[3vsv]] – TsXYL + xylose <BR /> | ||
| - | **[[3vsu]] – TsXYL + xylobiose <BR /> | ||
| - | }} | ||
==References== | ==References== | ||
<references /> | <references /> | ||
| + | </StructureSection> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||