Tachyplesin
From Proteopedia
(Difference between revisions)
| (59 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load=' | + | <StructureSection load='' size='340' side='right' caption='Tacyplesin I (PDB code [[1ma2]])' scene='67/671725/First_scene/2'> |
== Introduction == | == Introduction == | ||
| - | Tachyplesin I | + | '''Tachyplesin I, II and III''' are [http://en.wikipedia.org/wiki/Antimicrobial_peptides antimicrobial polypeptide] originally detected in the leukocytes of Japanese [http://en.wikipedia.org/wiki/Horseshoe_crab Horse Shoe Crab]. It has been reported to inhibit the growth of [http://en.wikipedia.org/wiki/Bacteria bacteria], [http://en.wikipedia.org/wiki/Fungus fungui] and [http://en.wikipedia.org/wiki/Virus viruses]. |
The antimicrobial activity of the polypeptide is contributed by electrostatic interaction between the negatively charged membrane of bacteria and fungi to positively charged part of <scene name='67/671725/Cationic_peptide_tpi/3'> TP-I </scene> <ref name=Laederach>PMID:12369825</ref> (see the {{Template:ColorKey_Hydrophobic}} and {{Template:ColorKey_Charge_Cationic}} amino acids). | The antimicrobial activity of the polypeptide is contributed by electrostatic interaction between the negatively charged membrane of bacteria and fungi to positively charged part of <scene name='67/671725/Cationic_peptide_tpi/3'> TP-I </scene> <ref name=Laederach>PMID:12369825</ref> (see the {{Template:ColorKey_Hydrophobic}} and {{Template:ColorKey_Charge_Cationic}} amino acids). | ||
Specifically, TP-I shows high affinity for negatively charged [http://en.wikipedia.org/wiki/Lipopolysaccharide lipopolysaccharides (LPS)] of [http://en.wikipedia.org/wiki/Gram-negative_bacteria gram-negative bacteria], thus neutralizing its effects. | Specifically, TP-I shows high affinity for negatively charged [http://en.wikipedia.org/wiki/Lipopolysaccharide lipopolysaccharides (LPS)] of [http://en.wikipedia.org/wiki/Gram-negative_bacteria gram-negative bacteria], thus neutralizing its effects. | ||
| Line 13: | Line 13: | ||
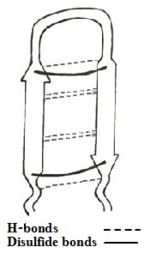
The sequence adapts an antiparallel β-sheet (hairpin) conformation in solution, with a <scene name='67/671725/Beta_turn_tp-1/2'>β-turn</scene> for the centrally located residues <scene name='67/671725/Tyrargglyile/3'>Tyr-Arg-Gly-Ile</scene>, stabilized by two cross-strand <scene name='67/671725/Disulfide_bonds/4'> disulfide bonds </scene> between Cys³-Cys¹⁶ and Cys⁷-Cys¹²<ref name=Saravanan>PMID:22464970</ref><ref name=Nakamura>Nakamura, Takanori, et al. "Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (''Tachypleus tridentatus''). Isolation and chemical structure." Journal of Biological Chemistry 263.32 (1988): 16709-16713</ref>, and [http://en.wikipedia.org/wiki/Protein_primary_structure C-terminus amidation]. In addition there are H-bonds and aromatic rings stacking interactions which helps stabilize the hairpin loop structure of the peptide. | The sequence adapts an antiparallel β-sheet (hairpin) conformation in solution, with a <scene name='67/671725/Beta_turn_tp-1/2'>β-turn</scene> for the centrally located residues <scene name='67/671725/Tyrargglyile/3'>Tyr-Arg-Gly-Ile</scene>, stabilized by two cross-strand <scene name='67/671725/Disulfide_bonds/4'> disulfide bonds </scene> between Cys³-Cys¹⁶ and Cys⁷-Cys¹²<ref name=Saravanan>PMID:22464970</ref><ref name=Nakamura>Nakamura, Takanori, et al. "Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (''Tachypleus tridentatus''). Isolation and chemical structure." Journal of Biological Chemistry 263.32 (1988): 16709-16713</ref>, and [http://en.wikipedia.org/wiki/Protein_primary_structure C-terminus amidation]. In addition there are H-bonds and aromatic rings stacking interactions which helps stabilize the hairpin loop structure of the peptide. | ||
| - | TP-I undergoes a conformational change | + | [http://en.wikipedia.org/wiki/Nuclear_magnetic_resonance NMR] studies have shown that TP-I undergoes a conformational change from <scene name='67/671725/First_scene/5'>water surrounding</scene> to <scene name='67/671725/Tp_i_in_the_presence_of_lps/4'>presence of LPS</scene>, making it <scene name='67/671725/Conformation_change/16'>more rigid and twisted</scene> than in the presence of water<ref name=Kushibiki>PMID:24389234</ref>. Moreover a docking model suggests the stability of the structure of TP-I is increased in the presence of LPS by the binding of the N and C termini of TP-I to LPS. The conformational change of TP-I seems to be crucial for its antimicrobial activity, since rearrangement of TP-I structure makes it more amphiphilic to negatively charged membrane of bacteria and fungus<ref name=Laederach>PMID:12369825</ref>. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== Derivatives or Analogue == | == Derivatives or Analogue == | ||
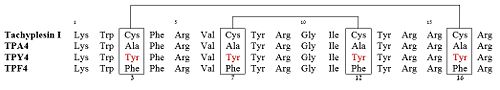
| - | Among all the existing interactions, cysteine | + | Among all the existing interactions, the cysteine bridges were considered as the principal contributors of the hairpin loop structure. To test this, three linear derivatives of TP-I (<scene name='67/671725/1ma4/3'>TPY4</scene>, TPF4 and TPA4) were created, in which the bridging cysteine residues were systematically replaced with tyrosine, phenylalanine, and alanine, respectively<ref name=Laederach>PMID:12369825</ref><ref name=Kushibiki>PMID:24389234</ref>. The linear derivatives of TP-I are mentioned below: |
| - | + | ||
| - | + | ||
| - | + | [[Image:Seq TPI.jpg|500px]] | |
| - | + | Of these 3 linear derivatives of TP-I, NMR structural investigations had shown that TPA4 was unstructured in solution. Also, TPA4 was inactive in terms of antimicrobial activity. In contrast, TPY4 and TPF4 adapt hairpin loop structure and also retain their antimicrobial properties, typical to TP-I. Therefore, the hairpin properties of the peptide seems to be important for recognition of LPS and its biological activities. | |
| - | + | ||
| - | CDT has been demonstrated to markedly inhibit the growth of | + | Besides replacement of cysteines, deletions were also performed in TP-I which yielded the surprising result of a hairpin loop that was seen, by NMR structure in LPS, in the <scene name='67/671725/Cdt/1'>Cysteine Deleted Tachyplesin</scene> (CDT). Thus, CTD with sequence NH₂-Lys-Trp-Phe-Arg-Val-Tyr-Arg-Gly-Ile-Tyr-Arg-Arg-Arg-CONH₂ did not have disulphide linkage, but was found to have broad spectrum of bactericidal activity. Specifically, CDT has been demonstrated to markedly inhibit the growth of [http://en.wikipedia.org/wiki/Escherichia_coli <i>Escherichia coli</i>] and [http://en.wikipedia.org/wiki/Listeria_monocytogenes <i>Listeria monocytogenes</i>] akin to TP-I, even with lower minimum inhibitory concentration (MIC) values. |
<b><u> CDT Structure </u></b> | <b><u> CDT Structure </u></b> | ||
| - | CDT, like TP-I, has a β-turn | + | CDT, like TP-I, has a β-turn with the <scene name='67/671725/Cdtturn/2'>same preserved residues</scene> in its LPS-bound structure. |
The β-hairpin topology of CDT is sustained by the <scene name='67/671725/Cdthaipin/2'>unique packing interactions</scene> between the aromatic ring of Trp2 and the side-chain of nonpolar amino acid of Val5 and the cationic side-chain of residue Arg11. | The β-hairpin topology of CDT is sustained by the <scene name='67/671725/Cdthaipin/2'>unique packing interactions</scene> between the aromatic ring of Trp2 and the side-chain of nonpolar amino acid of Val5 and the cationic side-chain of residue Arg11. | ||
| - | + | Also, there exists close proximity between residues <scene name='67/671725/Cdtnoe/1'>Trp2 and Ile9</scene> which is supported by [http://en.wikipedia.org/wiki/Nuclear_Overhauser_effect nuclear overhauser effects (NOEs)] involving [http://en.wikipedia.org/wiki/Indole indole] ring protons of Trp2 with side-chain proton of Ile9. These packing interactions have rendered an approximate anti-parallel orientation of the hairpin structure of CDT in presence of LPS. | |
| - | The β-hairpin | + | The β-hairpin structure displays an extended <font color='darkblue'>positively charged</font> surface patch of residues <scene name='67/671725/Cdtr4r7r12r13/3'>Arg 4, 7, 12 and 13</scene>. These positively charged basic residues interacts with the anionic phosphate groups of LPS, that leads to a plausible disruption or fluidization of LPS structures. This facilitates traversal of the peptide through the LPS-outer membrane<ref name=Saravanan>PMID:22464970</ref>. |
| - | + | ||
| - | + | ||
== Mode of action == | == Mode of action == | ||
| - | TP-I has affinity to LPS and also has ability to permeabilize the cell membrane of pathogens. TP-I | + | TP-I has high affinity to negatively charged cell membrane containing LPS and also has ability to permeabilize the cell membrane of pathogens. Docking model suggests strong interaction between cationic residues of TP-I with phosphate group and saccharides of LPS, where <scene name='67/671725/Conformation_change/9'> Arg 5 and Arg 14</scene>, acts as hinges <ref name=Laederach>PMID:12369825</ref>. Also, interaction between hydrophobic residues of TP-I with acyl chains of LPS was observed which strengthens the TP-I/LPS interaction<ref name=Hong>Hong, Jun, et al. "Mechanism of Tachyplesin I injury to bacterial membranes and intracellular enzymes, determined by laser confocal scanning microscopy and flow cytometry." Microbiological research (2014)</ref>. Ultimately, binding of TP-I/LPS neutralizes LPS, which is widely considered as endotoxin, and disrupts membrane function. |
| - | In addition to LPS binding, footpriting analysis has revealed the binding of TP-I to DNA by interacting specifically in minor groove of DNA duplex. The interaction between TP-I and DNA is contributed by secondary structure of the peptide which contains an antiparallel beta-sheet constrained by two disulfide bridges and connected by β-turn <ref name=Yonezawa>PMID:1372516</ref>. | + | In addition to LPS binding, footpriting analysis has revealed the binding of TP-I to DNA by interacting specifically in minor groove of DNA duplex. The interaction between TP-I and DNA is contributed by secondary structure of the peptide which contains an antiparallel beta-sheet constrained by two disulfide bridges and connected by β-turn <ref name=Yonezawa>PMID:1372516</ref>. TP-I on binding to DNA and RNA, inhibits the synthesis of macromolecules. |
| - | + | ||
| - | In summary, three processes might happen upon TP-I exposure: (1) Bacterial cell membranes are penetrated without disruption of the membrane and the peptide reaches the inner structures of the cell, damaging critical intracellular targets and interfering with intracellular functions and normal metabolism. (2) Pores are formed in the cell wall, causing leakage of intracellular content, leading to cell death. (3) DNA, RNA or protein synthesis are inhibited, killing the bacteria.<ref name=Hong>Hong, Jun, et al. "Mechanism of tachyplesin I injury to bacterial membranes and intracellular enzymes, determined by laser confocal scanning microscopy and flow cytometry." Microbiological research (2014) | + | In summary, three processes might happen upon TP-I exposure: (1) Bacterial cell membranes are penetrated without disruption of the membrane and the peptide reaches the inner structures of the cell, damaging critical intracellular targets and interfering with intracellular functions and normal metabolism. (2) Pores are formed in the cell wall, causing leakage of intracellular content, leading to cell death. (3) DNA, RNA or protein synthesis are inhibited, killing the bacteria.<ref name=Hong>Hong, Jun, et al. "Mechanism of tachyplesin I injury to bacterial membranes and intracellular enzymes, determined by laser confocal scanning microscopy and flow cytometry." Microbiological research (2014)</ref>. |
== Importance and relevance == | == Importance and relevance == | ||
| - | + | <b><u> Plants and Agriculture </u></b> | |
| - | + | Evidences suggest that TP-I has ability to permeabilize the cell membranes of pathogens.<ref name=Laederach>PMID:12369825</ref>. Also, LPS and DNA being the potential biological targets of the peptide, its antimicrobial activity might be exploited. Eyeing the potential of TP-I, it has been insetred successfully in genome of ''Ornithogalum dubium'' and ''Ornithogalum thyrsoides''. These ornamentals plants were originally sensitive to soft rot erwinias (SREs) and insertion of TP-I in the plants has successfully protected them without affecting their normal physiology <ref name=Lipsky>PMID:25438795</ref><ref name=Lipsky and Joshi>PMID:27639550</ref>. | |
| - | + | ||
| - | + | <b><u> Clinical Importance </u></b> | |
| - | + | Unlike mammalian cell membrane, bacterial cell membrane are negatively charged. [http://en.wikipedia.org/wiki/Escherichia_coli <i>Escherichia coli</i>] and [http://en.wikipedia.org/wiki/Staphylococcus_aureus <i>Listeria monocytogenes</i>] being most common pathogenic bacteria in animals and humans, studying the effect of TP-I on <i>E. coli</i> and <i>S. aureus</i> will be valuable in guiding clinical practice. The study in <i>E. coli</i> has shown membrane disruption upon treatment with TP-I. Also macromolecule leakage into the cytoplasm and the release of potassium ions was observed that ultimately killed <i>E. coli</i><ref name=Hong>Hong, Jun, et al. "Mechanism of tachyplesin I injury to bacterial membrane and intracellular enzymes, determined by laser confocal scanning microscopy and flow cytometry." Microbiological research (2014)</ref>. | |
| - | + | ||
== Possible Function as anti-tumor peptide == | == Possible Function as anti-tumor peptide == | ||
The cationic nature of Tachyplesin allows it to interact with anionic phospholipids present in the bacterial membrane and thereby disrupting membrane function. Besides this, the structural nature of Tachyplesin also highlights its antitumor properties. Since it can interact with the membrance of prokaryotic cell, it is likely that TP-I can also interact with the mitochondrial membrane of eukaryotic cells. Mitochondria are widely believed to have evolved from prokaryotic cells, that have established a symbiotic relationship with the primitive eukaryotic cell which signifies the structural similarity of mitochondrial and prokaryotic membranes. | The cationic nature of Tachyplesin allows it to interact with anionic phospholipids present in the bacterial membrane and thereby disrupting membrane function. Besides this, the structural nature of Tachyplesin also highlights its antitumor properties. Since it can interact with the membrance of prokaryotic cell, it is likely that TP-I can also interact with the mitochondrial membrane of eukaryotic cells. Mitochondria are widely believed to have evolved from prokaryotic cells, that have established a symbiotic relationship with the primitive eukaryotic cell which signifies the structural similarity of mitochondrial and prokaryotic membranes. | ||
| - | It was found that the synthetic Tachyplesin conjugated to the integrin homing domain (RGD-Tachyplesin) can inhibit the [http://en.wikipedia.org/wiki/Cell_growth proliferation] of TSU tumor cells [http://en.wikipedia.org/wiki/Prostate_cancer prostate cancer] and B16 [http://en.wikipedia.org/wiki/Melanoma melanoma] cells as well as [http://en.wikipedia.org/wiki/Endothelium endothelial cells] in a dose-dependent manner <i>in vitro</i> and reduce tumor growth <i>in vivo</i> by inducing [http://en.wikipedia.org/wiki/Apoptosis apoptosis].<ref name=Chen>PMID:11289111</ref>. Besides this RGD-Tachyplesin can activate caspases and induce Fas ligand, which are the markers for programmed cell death (PCD). Collectively, suppression of tumor associated cell and induction of programmed cell death will eventually act as therapy for cancer and tumor cells. | + | It was found that the synthetic Tachyplesin conjugated to the integrin homing domain (RGD-Tachyplesin) can inhibit the [http://en.wikipedia.org/wiki/Cell_growth proliferation] of TSU tumor cells [http://en.wikipedia.org/wiki/Prostate_cancer prostate cancer] and B16 [http://en.wikipedia.org/wiki/Melanoma melanoma] cells as well as [http://en.wikipedia.org/wiki/Endothelium endothelial cells] in a dose-dependent manner <i>in vitro</i> and reduce tumor growth <i>in vivo</i> by inducing [http://en.wikipedia.org/wiki/Apoptosis apoptosis].<ref name=Chen>PMID:11289111</ref>. Besides this RGD-Tachyplesin can activate caspases and induce Fas ligand, which are the markers for programmed cell death (PCD)<ref name=Ellrby>Ellerby, H. Michael, et al. "Anti-cancer activity of targeted pro-apoptotic peptides." Nature Medicine(1999)</ref>. |
| + | Collectively, suppression of tumor associated cell and induction of programmed cell death will eventually act as therapy for cancer and tumor cells. | ||
| + | ==3D structure of tachyplesin== | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| + | *Tachyplesin I | ||
| - | + | **[[1ma2]], [[1ma5]], [[1wo0]], [[1wo1]], [[2mdb]], [[2rtv]], [[6pin]] – TtTacI peptide residues 24-40 – Tachypleus tridentatus - - NMR<br /> | |
| - | *[[ | + | **[[1ma4]], [[1ma6]] – TtTacI peptide residues 24-40 (mutant) - NMR<br /> |
| + | **[[2lm8]] – TacI peptide residues 1-13 (mutant) - NMR<br /> | ||
| + | *Tachyplesin II | ||
| + | |||
| + | **[[6pio]] – TtTacII peptide residues 24-40 - NMR<br /> | ||
| + | **[[6pi2]] – TacII peptide residues 24-40 - Limulus polyphemus - NMR<br /> | ||
| + | |||
| + | *Tachyplesin III | ||
| + | |||
| + | **[[6pip]], [[6pi3]] – TacIII + peptide residues 24-40 - Tachypleus gigas - NMR<br /> | ||
| + | }} | ||
== Quiz == | == Quiz == | ||
| Line 89: | Line 87: | ||
{What is the secondery structure of TP-I?} | {What is the secondery structure of TP-I?} | ||
| - | + Two antiparallel β- | + | + Two antiparallel β-strands |
- Two antiparallel α-Helixes | - Two antiparallel α-Helixes | ||
| - | - Two parallel β- | + | - Two parallel β-strands |
- Two parallel α-Helixes | - Two parallel α-Helixes | ||
| Line 97: | Line 95: | ||
- TPF4 | - TPF4 | ||
- TPY4 | - TPY4 | ||
| - | + | + TPA4 | |
- CDT | - CDT | ||
| Line 113: | Line 111: | ||
</Quiz> | </Quiz> | ||
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | </StructureSection> | ||
| + | [[Category:Topic Page]] | ||
| + | [[Category:Pages with quizzes]] | ||
Current revision
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Shulamit Idzikowski, Janak Raj Joshi, Michal Harel, Alexander Berchansky, Joel L. Sussman, Angel Herraez, Jaime Prilusky