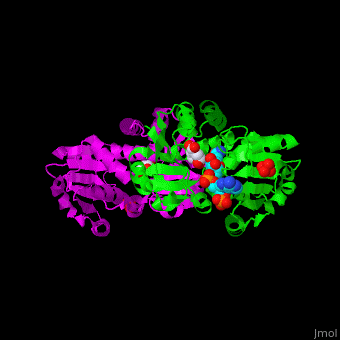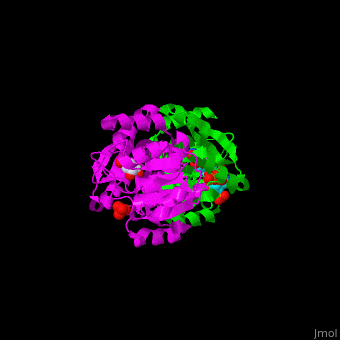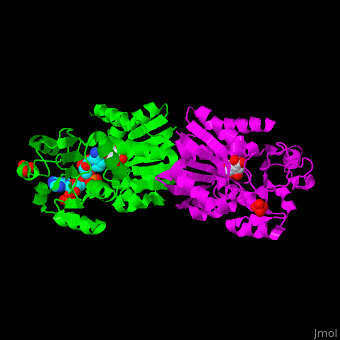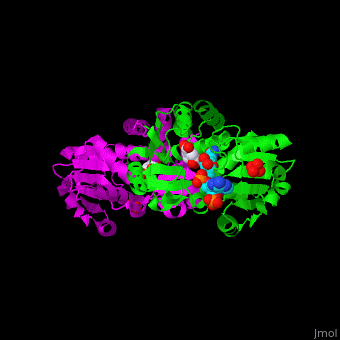
Shikimate dehydrogenase
From Proteopedia
(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
<StructureSection load='' size='350' side='right' scene='54/540162/Cv/1' caption='Shikimate dehydrogenase dimer complex with shikimate, sulfate and NADP [[2ev9]]'> | <StructureSection load='' size='350' side='right' scene='54/540162/Cv/1' caption='Shikimate dehydrogenase dimer complex with shikimate, sulfate and NADP [[2ev9]]'> | ||
| + | __TOC__ | ||
== Function == | == Function == | ||
'''Shikimate dehydrogenase''' (AroE) catalyzes the conversion of shikimate and NADP+ to 3-dihydroshikimate, NADPH and H+. It is part of the shikimate pathway which is responsible for the biosynthesis of phenylalanine, tyrosine and tryptophan. This pathway is found in bacteria, fungi, plants, algae and parasites and is missing in animals and humans<ref>PMID:18669580</ref>. | '''Shikimate dehydrogenase''' (AroE) catalyzes the conversion of shikimate and NADP+ to 3-dihydroshikimate, NADPH and H+. It is part of the shikimate pathway which is responsible for the biosynthesis of phenylalanine, tyrosine and tryptophan. This pathway is found in bacteria, fungi, plants, algae and parasites and is missing in animals and humans<ref>PMID:18669580</ref>. | ||
| Line 5: | Line 6: | ||
==Structural highlights == | ==Structural highlights == | ||
AroE changes its conformation from an open to closed one upon binding of the cofactor NADP. The active site contains the substrate shikimate<ref>PMID:17825835</ref>. | AroE changes its conformation from an open to closed one upon binding of the cofactor NADP. The active site contains the substrate shikimate<ref>PMID:17825835</ref>. | ||
| - | *<scene name='54/540162/Cv/ | + | *<scene name='54/540162/Cv/5'>NADP binding site</scene>. Water molecules are shown as red spheres. |
| - | *<scene name='54/540162/Cv/ | + | *<scene name='54/540162/Cv/6'>Whole binding site</scene>. |
| - | + | ||
== 3D Structures of shikimate dehydrogenase == | == 3D Structures of shikimate dehydrogenase == | ||
| - | + | [[Shikimate dehydrogenase 3D structures]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== References == | == References == | ||
| + | </StructureSection> | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
- ↑ Singh S, Stavrinides J, Christendat D, Guttman DS. A phylogenomic analysis of the shikimate dehydrogenases reveals broadscale functional diversification and identifies one functionally distinct subclass. Mol Biol Evol. 2008 Oct;25(10):2221-32. doi: 10.1093/molbev/msn170. Epub 2008 Jul, 31. PMID:18669580 doi:http://dx.doi.org/10.1093/molbev/msn170
- ↑ Bagautdinov B, Kunishima N. Crystal structures of shikimate dehydrogenase AroE from Thermus thermophilus HB8 and its cofactor and substrate complexes: insights into the enzymatic mechanism. J Mol Biol. 2007 Oct 19;373(2):424-38. Epub 2007 Aug 21. PMID:17825835 doi:10.1016/j.jmb.2007.08.017
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Sohail Jooma