Journal:Acta Cryst F:S2053230X20010122
From Proteopedia
(Difference between revisions)

| (3 intermediate revisions not shown.) | |||
| Line 5: | Line 5: | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
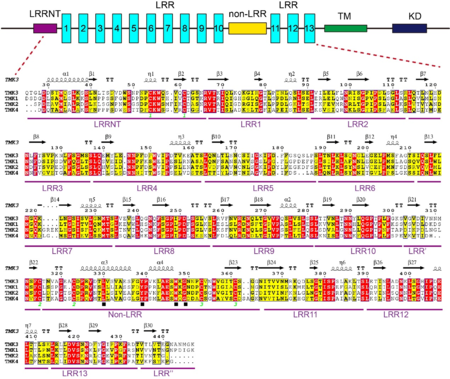
In this study, we successfully expressed the extracellular domain of TMK3-ECD (residues 25~482) in an insect-cell secretion expression system. We obtained diffraction-quality crystals and determined the three-dimensional structure for TMK3-ECD. The structure was refined to a resolution of 2.06 Å with R<sub>''work''</sub> of 17.69% and R<sub>''free''</sub> of 20.58% (Table 1). Except for 32 residues from the C-terminus and Gln25 from the N-terminus, all remaining residues were well defined and included in the final refined model. The overall structure of the TMK3-LRR exhibits a “L” shape: | In this study, we successfully expressed the extracellular domain of TMK3-ECD (residues 25~482) in an insect-cell secretion expression system. We obtained diffraction-quality crystals and determined the three-dimensional structure for TMK3-ECD. The structure was refined to a resolution of 2.06 Å with R<sub>''work''</sub> of 17.69% and R<sub>''free''</sub> of 20.58% (Table 1). Except for 32 residues from the C-terminus and Gln25 from the N-terminus, all remaining residues were well defined and included in the final refined model. The overall structure of the TMK3-LRR exhibits a “L” shape: | ||
| - | *<scene name='85/857139/Cv/10'>Overall structure of TMK3-LRR domain. 1st orientation</scene>. | + | *<scene name='85/857139/Cv/10'>Overall structure of TMK3-LRR domain. 1st orientation</scene> (PDB entry [[7brc]]). |
*<scene name='85/857139/Cv/11'>Overall structure of TMK3-LRR domain. 2nd orientation</scene>. The LRRNT, LRRs (numbered as indicated) and the non-LRR region of TMK3-LRR are colored in green, whitesmoke and blue, respectively. The three disulfide bonds (Cys54-Cys61, Cys315-Cys323 and Cys353-Cys361) are depicted in yellow. “N” and “C” represent N- and C-terminus, respectively. | *<scene name='85/857139/Cv/11'>Overall structure of TMK3-LRR domain. 2nd orientation</scene>. The LRRNT, LRRs (numbered as indicated) and the non-LRR region of TMK3-LRR are colored in green, whitesmoke and blue, respectively. The three disulfide bonds (Cys54-Cys61, Cys315-Cys323 and Cys353-Cys361) are depicted in yellow. “N” and “C” represent N- and C-terminus, respectively. | ||
A block of about 40 residues termed non-LRR region intersects the LRR domain into two subdomains – a N-terminal LRR subdomain with 10 LRRs (N-LRRs) and a C-terminal LRR subdomain with 3 LRRs (C-LRRs). C-LRR packs nearly perpendicularly against the spine of N-LRR. Most LRRs from the N-LRR contain the sequence GxL/i/vP (x stands for any amino acid) specific for the plant LRR proteins, with the only exceptions for LRR3, LRR4, LRR7 and LRR10 (see static image below). | A block of about 40 residues termed non-LRR region intersects the LRR domain into two subdomains – a N-terminal LRR subdomain with 10 LRRs (N-LRRs) and a C-terminal LRR subdomain with 3 LRRs (C-LRRs). C-LRR packs nearly perpendicularly against the spine of N-LRR. Most LRRs from the N-LRR contain the sequence GxL/i/vP (x stands for any amino acid) specific for the plant LRR proteins, with the only exceptions for LRR3, LRR4, LRR7 and LRR10 (see static image below). | ||
| Line 21: | Line 21: | ||
{{Clear}} | {{Clear}} | ||
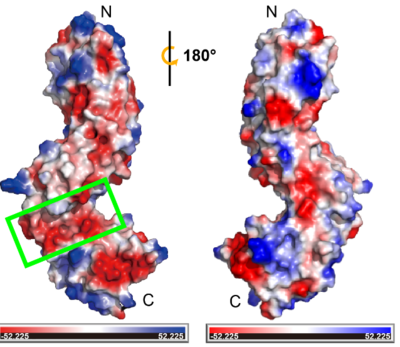
This suggests that the non-LRR region is likely a ligand binding pocket that can accommodate positively charged ligands, such as peptides with basic side chains or other electropositive signaling molecules. In addition, the disulfide bonds that stabilize and orient the fold of non-LRR region in TMK3-LRR could be essential for functionality. The disulfide bond between Cys353 to Cys361 is conserved with classical N-cap structure, suggesting its function as an N-cap to stabilize the structure of C-LRR. The other disulfide bond (Cys315-Cys323) is responsible for connecting two LRR domains. Breaking this pairs of disulfide bond may cause the two LRR domains to be converted into one, altering the ligand binding interaction. Besides, it may also play a critical role in a changing redox environment. The number of N-LRR is greater than the number of C-LRR, which brings close proximity of the non-LRR to interact with ligand and the transmembrane region. It also might facilitate signaling in the cytoplasm through interactions with ligand. We also speculate it's possible to have a role for the N- LRR and C-LRR domains that provides more than one scaffold for protein interaction or ligand recognition. | This suggests that the non-LRR region is likely a ligand binding pocket that can accommodate positively charged ligands, such as peptides with basic side chains or other electropositive signaling molecules. In addition, the disulfide bonds that stabilize and orient the fold of non-LRR region in TMK3-LRR could be essential for functionality. The disulfide bond between Cys353 to Cys361 is conserved with classical N-cap structure, suggesting its function as an N-cap to stabilize the structure of C-LRR. The other disulfide bond (Cys315-Cys323) is responsible for connecting two LRR domains. Breaking this pairs of disulfide bond may cause the two LRR domains to be converted into one, altering the ligand binding interaction. Besides, it may also play a critical role in a changing redox environment. The number of N-LRR is greater than the number of C-LRR, which brings close proximity of the non-LRR to interact with ligand and the transmembrane region. It also might facilitate signaling in the cytoplasm through interactions with ligand. We also speculate it's possible to have a role for the N- LRR and C-LRR domains that provides more than one scaffold for protein interaction or ligand recognition. | ||
| + | |||
| + | <scene name='85/857139/Cv/13'>TMK3-LRR structure alignment of the TMK1-LRR are shown in blue (TMK1) and red (TMK3)</scene>. | ||
| + | |||
| + | <scene name='85/857139/Cv/14'>Alignment with TMK3-LRR, BRI1-LRR and PSYR-LRR</scene>. The structure of the TMK3-LRR, BRI1-LRR and PSYR-LRR are depicted in red, blue and yellow. | ||
| + | |||
| + | '''PDB reference:''' extracellular domain of TMK3, [[7brc]]. | ||
<b>References</b><br> | <b>References</b><br> | ||
Current revision
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.