SARS-CoV-2 protein NSP11
From Proteopedia
(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
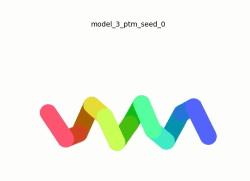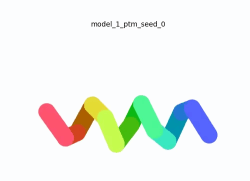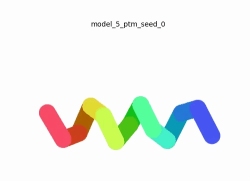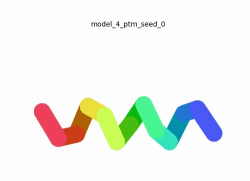
To the right is an '''[[AlphaFold]]2''' 3D model of SARS CoV-2 Protein NSP11 (length = 13 amino acids, i.e. '''SADAQSFLNGFAV''' color coded by the pLDDT scores. It corresponds to the highest ranked model in terms of the pLDDT confidence scores, ''i.e.'', model 5<ref name="MIT_ColabFold">[https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/beta/AlphaFold2_advanced.ipynb MIT ColabFold]</ref>. | To the right is an '''[[AlphaFold]]2''' 3D model of SARS CoV-2 Protein NSP11 (length = 13 amino acids, i.e. '''SADAQSFLNGFAV''' color coded by the pLDDT scores. It corresponds to the highest ranked model in terms of the pLDDT confidence scores, ''i.e.'', model 5<ref name="MIT_ColabFold">[https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/beta/AlphaFold2_advanced.ipynb MIT ColabFold]</ref>. | ||
[[Image:NSP11_AlphaFold2_Movie.gif|250px|left|thumb|'''Morph''' of the top 5 ranked [[AlphaFold]]2 models of '''SARS-CoV-2 Protein NSP11''', ''rainbow'' color coded N-C (blue to red)<ref name="MIT_ColabFold"/>.]] | [[Image:NSP11_AlphaFold2_Movie.gif|250px|left|thumb|'''Morph''' of the top 5 ranked [[AlphaFold]]2 models of '''SARS-CoV-2 Protein NSP11''', ''rainbow'' color coded N-C (blue to red)<ref name="MIT_ColabFold"/>.]] | ||
| + | |||
| + | |||
== Function == | == Function == | ||
| - | '''Non-structural protein 11 (NSP11)''' is the | + | '''Non-structural protein 11 (NSP11)''' is the short peptide at the C-term of the large SARS-CoV-2 polyprotein ORF1ab (pp1a). it is the cleavage product of pp1a polyprotein by 3CLpro/Mpro protease at the nsp10/11 junction. NSP11 overlaps a portion of the RNA that codes for NSP12, so whether NSP11 is actually expressed as well as its putative function is not known<ref name="NY Times COVID-19">[https://www.nytimes.com/interactive/2020/04/03/science/coronavirus-genome-bad-news-wrapped-in-protein.html ''NY Times'' (3-Apr-2020) Bad News Wrapped in Protein: Inside the Coronavirus Genome]</ref>. |
| Line 10: | Line 12: | ||
Giri's lab<ref>PMID: 34119626</ref> studied NSP11 in solution and by molecular dynamics. Based on CD, they found that NSP11 shows characteristics of an intrinsically disordered protein (IDP). They further studied the its conformational behavior in the presence of negatively charged and neutral liposomes, and found that it remains disordered. However, in SDS micelle, NSP11 adopts an α-helical conformation, suggesting the helical propensity of NSP11, which is very similar to what [AlphaFold[2 predicts. Based on MD simulations NSP11 appears to to have the behavior of an IDP. | Giri's lab<ref>PMID: 34119626</ref> studied NSP11 in solution and by molecular dynamics. Based on CD, they found that NSP11 shows characteristics of an intrinsically disordered protein (IDP). They further studied the its conformational behavior in the presence of negatively charged and neutral liposomes, and found that it remains disordered. However, in SDS micelle, NSP11 adopts an α-helical conformation, suggesting the helical propensity of NSP11, which is very similar to what [AlphaFold[2 predicts. Based on MD simulations NSP11 appears to to have the behavior of an IDP. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
== See also == | == See also == | ||
| - | [[Coronavirus_Disease 2019 (COVID-19)]] | + | [[Coronavirus_Disease 2019 (COVID-19)]]<br> |
| + | [[SARS-CoV-2_virus_proteins]]<br> | ||
| + | [[COVID-19 AlphaFold2 Models]] | ||
__NOTOC__ | __NOTOC__ | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 MIT ColabFold
- ↑ NY Times (3-Apr-2020) Bad News Wrapped in Protein: Inside the Coronavirus Genome
- ↑ Gadhave K, Kumar P, Kumar A, Bhardwaj T, Garg N, Giri R. Conformational dynamics of 13 amino acids long NSP11 of SARS-CoV-2 under membrane mimetics and different solvent conditions. Microb Pathog. 2021 Sep;158:105041. doi: 10.1016/j.micpath.2021.105041. Epub 2021, Jun 10. PMID:34119626 doi:http://dx.doi.org/10.1016/j.micpath.2021.105041