User:Charli Barbet/Sandbox
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
'''Grb2 (1gri)''' | '''Grb2 (1gri)''' | ||
<StructureSection load='1gri' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='1gri' size='340' side='right' caption='Caption for this structure' scene=''> | ||
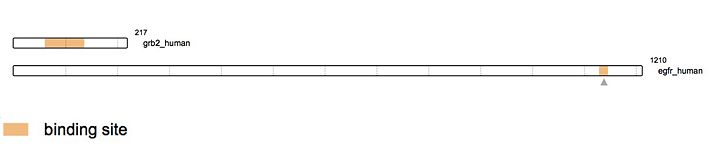
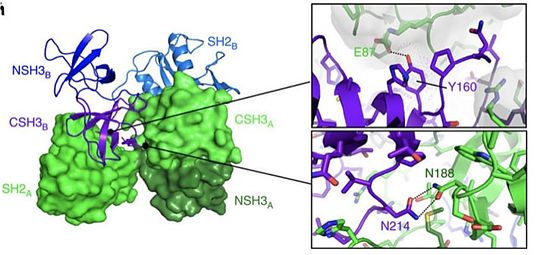
| - | Growth Factor Receptor Bound Protein is a cytosolic protein made of 217 amino acids and weighing 25,206 Da. Ubiquitously present in the cell, | + | Growth Factor Receptor Bound Protein (Grb2) is a cytosolic protein made of 217 amino acids and weighing 25,206 Da. Ubiquitously present in the cell, the protein is involved in signal transduction and especially in the [https://en.wikipedia.org/wiki/MAPK/ERK_pathway MAP kinase pathway]. Grb2 interacts mainly with [https://en.wikipedia.org/wiki/Tyrosine_kinase tyrosine kinases] such as [http://www.uniprot.org/uniprot/P00533 EGFR] once the latest has been activated by ligand binding. This specific binding leads to the recruitment of [https://en.wikipedia.org/wiki/Guanine_nucleotide_exchange_factor GEF] (like [http://www.uniprot.org/uniprot/Q07889 SOS1]), stimulating the activation of other pathways. Several others interactions have been elucidated like the capacity of the protein to dimerise thus proving its potential implication in the growth of [https://en.wikipedia.org/wiki/Malignancy malignant cells]. |
== Structure == | == Structure == | ||
| Line 13: | Line 13: | ||
<scene name='75/750264/Sh2/1'>SH2 DOMAIN</scene>: | <scene name='75/750264/Sh2/1'>SH2 DOMAIN</scene>: | ||
| - | SH2 domain is a domain that is approximately 100 amino acids long and with a very conserved structure. Identified | + | SH2 domain is a domain that is approximately 100 amino acids long and with a very conserved structure. Identified in several human and rodent proteins such as [https://en.wikipedia.org/wiki/Phosphatase phosphatases], [https://en.wikipedia.org/wiki/Transcription_factor transcription factor], or [https://en.wikipedia.org/wiki/Signal_transducing_adaptor_protein adaptor] like protein such as Grb2 for instance. This domain is therefore ubiquitous in several cellular signaling pathways.Typically, the SH2 domain specifically recognizes sites with phosphorylated tyrosine in different types of proteins. SH2 can for instance bind to the intracellular region of EGF leading in turn, to the formation of protein signalization complexes. This binding and the role of SH2 is very important in the conversion of an extra-cellular signal in an intra-cellular signal giving rise to diversified cellular responses or the expression of specific genes.It is also important to note that the SH2 domain can bind to other SH2 domains.However, a mutation in the specific binding site of SH2 can impede the interaction of two proteins and thus the formation of a protein complex. Therefore, mutations in SH2 can give rise to cellular dysfunction and lead to several diseases. |
<scene name='75/750264/Sh3/1'>SH3 DOMAIN</scene>: | <scene name='75/750264/Sh3/1'>SH3 DOMAIN</scene>: | ||
| - | The SH3 domain is a region of a protein that is approximately 50 amino acid long. Largely | + | The SH3 domain is a region of a protein that is approximately 50 amino acid long. Largely expressed in proteins associated to the membrane. The domain is made of 5 to 6 β-sheets arranged in two antiparallel β-sheets. The linking region between the two β-sheets can contain α helices. This special conformation allows the arrangement of a hydrophobic pocket in which the ligand can bind. Typically, the binding region has a motif rich in Prolines: PXXP. This binding allows the formation of multi-proteins complexes involved in the translation of an extra-cellular signal and its conversion. The binding is thus largely involved in gene expression and protein concentration. |
ISOFORM: | ISOFORM: | ||
| - | + | Grb2 posses an isoform, known as Grb3.3. | |
| + | Grb3.3 is present in cells but it induces apoptosis. This isoform has a very similar structure to Grb2 but is truncated from an SH3 domain (from the 60th to the 100th amino) resulting in a degradation of its SH2 domain and a loss of functionality. | ||
== Function == | == Function == | ||
| - | The Grb2 isoform has a non-functional SH2 domain, unable to | + | The Grb2 isoform has a non-functional SH2 domain, unable to bind the phosphorylated tyrosine of its targeted protein (EGFR for instance). The inability of the molecule to transmit signal is translated by apoptosis of the cell, thus regulating growth signal. |
| - | The functional isoform Grb2 is involved in several cellular functions detailed below | + | The functional isoform: Grb2, is involved in several cellular functions detailed below: |
| + | |||
| + | On one hand, the SH2 domain recognizes phosphorylated residues which are mainly tyrosines. The recognized tyrosines present a caracteristic motif for recognition: NH2-pYXNX-COOH. | ||
- pY representing the phosphorylated tyrosine. | - pY representing the phosphorylated tyrosine. | ||
| Line 31: | Line 34: | ||
Thus by the special recognition of this motif, the binding of the two molecules is very specific. These motifs are highly expressed in several cellular proteins like [https://en.wikipedia.org/wiki/Receptor_tyrosine_kinase Receptor Tyrosine Kinase] ([http://www.uniprot.org/uniprot/P00533 epidermal growth factor receptor], [http://www.uniprot.org/uniprot/P11362 fibroblast growth factor receptor]) but equally in proteins that are not [https://en.wikipedia.org/wiki/Receptor_tyrosine_kinase Receptor Tyrosine Kinase] ([http://www.uniprot.org/uniprot/Q05397 focal adhesion kinase], [http://www.uniprot.org/uniprot/P35568 insulin receptor substrate-1]). | Thus by the special recognition of this motif, the binding of the two molecules is very specific. These motifs are highly expressed in several cellular proteins like [https://en.wikipedia.org/wiki/Receptor_tyrosine_kinase Receptor Tyrosine Kinase] ([http://www.uniprot.org/uniprot/P00533 epidermal growth factor receptor], [http://www.uniprot.org/uniprot/P11362 fibroblast growth factor receptor]) but equally in proteins that are not [https://en.wikipedia.org/wiki/Receptor_tyrosine_kinase Receptor Tyrosine Kinase] ([http://www.uniprot.org/uniprot/Q05397 focal adhesion kinase], [http://www.uniprot.org/uniprot/P35568 insulin receptor substrate-1]). | ||
| - | As an example, the SH2 domain of Grb2 recognizes an intracellular phosphorylated tyrosine. This binding | + | As an example, the SH2 domain of Grb2 recognizes an intracellular phosphorylated tyrosine. This binding leads to the recruitment of [http://www.uniprot.org/uniprot/Q07889 SOS-1] via the SH3 domain of Grb2. Indeed, Grb2 is also made of two SH3 domains. These domains are able to recognize Proline rich region like the one of [http://www.uniprot.org/uniprot/Q07889 SOS-1] protein (Son Of Sevenless). |
Following this pathway and the formation of a complex between Grb2 and [http://www.uniprot.org/uniprot/Q07889 SOS], the [http://www.uniprot.org/uniprot/P01112 RAS] protein is activated. Interestingly, [http://www.uniprot.org/uniprot/P01112 RAS] is a g-protein implicated in the activation of [http://www.uniprot.org/uniprot/P04049 RAF-1]. The latest activates of the [https://en.wikipedia.org/wiki/MAPK/ERK_pathway MEK downstream cascade pathway] ([http://www.uniprot.org/uniprot/Q02750 MEK1]/ [http://www.uniprot.org/uniprot/P36507 MEK2] et [http://www.uniprot.org/uniprot/P27361 ERK1 ]/ [http://www.uniprot.org/uniprot/P28482 ERK2]) involved in the translocation of [https://en.wikipedia.org/wiki/Extracellular_signal–regulated_kinases ERK factors] from the cytosol to the nucleus for the activation of [http://www.uniprot.org/uniprot/P19419 Elk-1] and [http://www.uniprot.org/uniprot/P01106 Myc transcription Factor]. These particular [https://en.wikipedia.org/wiki/Transcription_factor transcription factor] participate in the activation of SRE containing gene leading to cellular growth. | Following this pathway and the formation of a complex between Grb2 and [http://www.uniprot.org/uniprot/Q07889 SOS], the [http://www.uniprot.org/uniprot/P01112 RAS] protein is activated. Interestingly, [http://www.uniprot.org/uniprot/P01112 RAS] is a g-protein implicated in the activation of [http://www.uniprot.org/uniprot/P04049 RAF-1]. The latest activates of the [https://en.wikipedia.org/wiki/MAPK/ERK_pathway MEK downstream cascade pathway] ([http://www.uniprot.org/uniprot/Q02750 MEK1]/ [http://www.uniprot.org/uniprot/P36507 MEK2] et [http://www.uniprot.org/uniprot/P27361 ERK1 ]/ [http://www.uniprot.org/uniprot/P28482 ERK2]) involved in the translocation of [https://en.wikipedia.org/wiki/Extracellular_signal–regulated_kinases ERK factors] from the cytosol to the nucleus for the activation of [http://www.uniprot.org/uniprot/P19419 Elk-1] and [http://www.uniprot.org/uniprot/P01106 Myc transcription Factor]. These particular [https://en.wikipedia.org/wiki/Transcription_factor transcription factor] participate in the activation of SRE containing gene leading to cellular growth. | ||
Revision as of 11:21, 15 January 2017
Grb2 (1gri)
| |||||||||||