Carboxypeptidase A
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
===Active Site=== | ===Active Site=== | ||
| - | The active site of bovine pancreatic CPA is embedded within a <scene name='69/694222/3cpadeeppocket/1'>deep pocket</scene> (colored orange) whose opening is located on the surface of the protein. When no polypeptide substrate is bound in the active site, the pocket is open. However, the pocket is "capped" by a tyrosine residue (Tyr248) (see section titled "Important Tyr248 Residue") when a substrate or [http://en.wikipedia.org/wiki/Enzyme_inhibitor inhibitor] molecule binds.<ref name="CPA2" /> Kinetics experiments have indicated that the binding region of the active site is actually capable of extending over five amino acids of the substrate.<ref name="CPA2" /> The active site contains two separate subsites, labeled S1' and S1, which each contain several pertinent residues that serve important roles during the catalyzed hydrolysis reaction. | + | The active site of bovine pancreatic CPA is embedded within a <scene name='69/694222/3cpadeeppocket/1'>deep pocket</scene> (colored orange) whose opening is located on the surface of the protein. When no polypeptide substrate is bound in the active site, the pocket is open. However, the pocket is <scene name='69/694222/3cpadeeppocket2/1'>"capped" by a tyrosine residue (Tyr248)</scene> (see section titled "Important Tyr248 Residue") when a substrate or [http://en.wikipedia.org/wiki/Enzyme_inhibitor inhibitor] molecule binds.<ref name="CPA2" /> In this view, the Tyr248 is shown in green. Kinetics experiments have indicated that the binding region of the active site is actually capable of extending over five amino acids of the substrate.<ref name="CPA2" /> The active site contains two separate subsites, labeled S1' and S1, which each contain several pertinent residues that serve important roles during the catalyzed hydrolysis reaction. |
=====S1' Subsite: The Hydrophobic Binding Pocket===== | =====S1' Subsite: The Hydrophobic Binding Pocket===== | ||
| Line 26: | Line 26: | ||
==Important Tyr248 Residue== | ==Important Tyr248 Residue== | ||
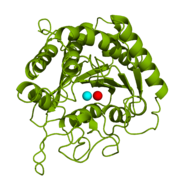
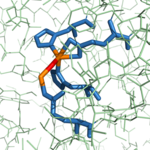
CPA from ''B. taurus'' has been co-crystallized with two Zn<sup>2+</sup> ions (Figure 1). This structure has been deposited in the PDB database under the label [http://www.rcsb.org/pdb/explore/explore.do?structureId=1cpx 1CPX], which is a β-form of CPA. 1CPX has an interesting distinction from some other crystallized CPA proteins in that its crystallographic data has revealed a different conformation of the Tyr248 residue.[[Image:1CPX-Tyr248.png|thumb|Figure 2: Important Tyr248 residue of 1CPX is pointing toward the hydrophobic binding pocket.]]<ref name="CPA1">Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. ''Biochemistry''. 37(47):16555-16564. [http://pubs.acs.org/doi/abs/10.1021/bi981678i DOI: 10.1021/bi981678i]</ref> Previous literature has suggested that the conserved Tyrosine residue among CPA proteins has been involved in an [http://en.wikipedia.org/wiki/Enzyme_catalysis#Induced_fit induced fit mechanism] because Tyr248 typically has been found <scene name='69/694222/Tyr248_5cpa/1'>pointing outward</scene> toward solution when a substrate is not bound in the active site.<ref name="CPA1">Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. ''Biochemistry''. 37(47):16555-16564. [http://pubs.acs.org/doi/abs/10.1021/bi981678i DOI: 10.1021/bi981678i]</ref> However, crystallographic data for 1CPX shows Tyr248 <scene name='69/694222/Tyr248_1cpx/1'>pointing toward the active site</scene> (Figure 2) without a polypeptide substrate bound. Therefore, this conflicts with the previously proposed induced fit mechanism for CPA proteins and suggests that Tyr248 is liganded to the catalytic Zn<sup>2+</sup> ion in 1CPX.<ref name="CPA1">Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. ''Biochemistry''. 37(47):16555-16564. [http://pubs.acs.org/doi/abs/10.1021/bi981678i DOI: 10.1021/bi981678i]</ref> Moreover, this data supports that this is the native conformation of Tyr248 in solution because none of the residues in the protein’s active site interact in crystallographic packing.<ref name="CPA1">Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. ''Biochemistry''. 37(47):16555-16564. [http://pubs.acs.org/doi/abs/10.1021/bi981678i DOI: 10.1021/bi981678i]</ref> Not only does Tyr248 point toward the active site, but in doing so, Tyr248 <scene name='69/694222/Tyr248_pointing_toward/2'>caps the hydrophobic binding pocket</scene> (shown in blue) with its interactions with Ile247 and a hydrogen bond.<ref name="CPA1">Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. ''Biochemistry''. 37(47):16555-16564. [http://pubs.acs.org/doi/abs/10.1021/bi981678i DOI: 10.1021/bi981678i]</ref> This point of view is able to clearly show how Tyr248 is thought to be a ligand with the catalytic Zn<sup>2+</sup> ion. | CPA from ''B. taurus'' has been co-crystallized with two Zn<sup>2+</sup> ions (Figure 1). This structure has been deposited in the PDB database under the label [http://www.rcsb.org/pdb/explore/explore.do?structureId=1cpx 1CPX], which is a β-form of CPA. 1CPX has an interesting distinction from some other crystallized CPA proteins in that its crystallographic data has revealed a different conformation of the Tyr248 residue.[[Image:1CPX-Tyr248.png|thumb|Figure 2: Important Tyr248 residue of 1CPX is pointing toward the hydrophobic binding pocket.]]<ref name="CPA1">Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. ''Biochemistry''. 37(47):16555-16564. [http://pubs.acs.org/doi/abs/10.1021/bi981678i DOI: 10.1021/bi981678i]</ref> Previous literature has suggested that the conserved Tyrosine residue among CPA proteins has been involved in an [http://en.wikipedia.org/wiki/Enzyme_catalysis#Induced_fit induced fit mechanism] because Tyr248 typically has been found <scene name='69/694222/Tyr248_5cpa/1'>pointing outward</scene> toward solution when a substrate is not bound in the active site.<ref name="CPA1">Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. ''Biochemistry''. 37(47):16555-16564. [http://pubs.acs.org/doi/abs/10.1021/bi981678i DOI: 10.1021/bi981678i]</ref> However, crystallographic data for 1CPX shows Tyr248 <scene name='69/694222/Tyr248_1cpx/1'>pointing toward the active site</scene> (Figure 2) without a polypeptide substrate bound. Therefore, this conflicts with the previously proposed induced fit mechanism for CPA proteins and suggests that Tyr248 is liganded to the catalytic Zn<sup>2+</sup> ion in 1CPX.<ref name="CPA1">Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. ''Biochemistry''. 37(47):16555-16564. [http://pubs.acs.org/doi/abs/10.1021/bi981678i DOI: 10.1021/bi981678i]</ref> Moreover, this data supports that this is the native conformation of Tyr248 in solution because none of the residues in the protein’s active site interact in crystallographic packing.<ref name="CPA1">Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. ''Biochemistry''. 37(47):16555-16564. [http://pubs.acs.org/doi/abs/10.1021/bi981678i DOI: 10.1021/bi981678i]</ref> Not only does Tyr248 point toward the active site, but in doing so, Tyr248 <scene name='69/694222/Tyr248_pointing_toward/2'>caps the hydrophobic binding pocket</scene> (shown in blue) with its interactions with Ile247 and a hydrogen bond.<ref name="CPA1">Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. ''Biochemistry''. 37(47):16555-16564. [http://pubs.acs.org/doi/abs/10.1021/bi981678i DOI: 10.1021/bi981678i]</ref> This point of view is able to clearly show how Tyr248 is thought to be a ligand with the catalytic Zn<sup>2+</sup> ion. | ||
| - | |||
| - | For a space-fill view of Tyr248 capping the binding pocket, <scene name='69/694222/3cpadeeppocket2/1'>click here</scene>. In this view, the Tyr248 is shown in green, while the binding pocket is displayed in orange. | ||
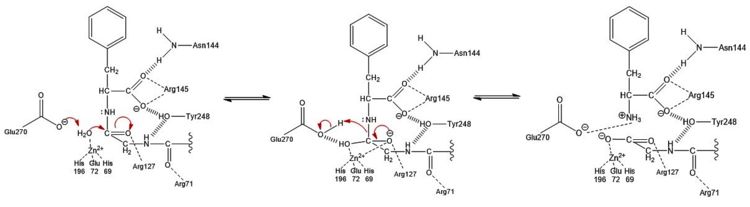
== Mechanism of Action == | == Mechanism of Action == | ||
Revision as of 13:46, 11 April 2017
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Carboxypeptidase A in Bos taurus
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. Biochemistry. 37(47):16555-16564. DOI: 10.1021/bi981678i
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Christianson DW, Lipscomb WN. 1989. Carboxypeptidase A. Acc. Chem. Res. 22:62-69.
- ↑ Suh J, Cho W, Chung S. 1985. Carboxypeptidase A-catalyzed hydrolysis of α-(acylamino)cinnamoyl derivatives of L-β-phenyllactate and L-phenylalaninate: evidence for acyl-enzyme intermediates. J. Am. Chem. Soc. 107:4530-4535. DOI: 10.1021/ja00301a025
- ↑ Geoghegan, KF, Galdes, A, Martinelli, RA, Holmquist, B, Auld, DS, Vallee, BL. 1983. Cryospectroscopy of intermediates in the mechanism of carboxypeptidase A. Biochem. 22(9):2255-2262. DOI: 10.1021/bi00278a031
- ↑ Kaplan, AP, Bartlett, PA. 1991. Synthesis and evaluation of an inhibitor of carboxypeptidase A with a Ki value in the femtomolar range. Biochem. 30(33):8165-8170. PMID: 1868091
- ↑ Worthington, K., Worthington, V. 1993. Worthington Enzyme Manual: Enzymes and Related Biochemicals. Freehold (NJ): Worthington Biochemical Corporation; [2011; accessed March 28, 2017]. Carboxypeptidase A. http://www.worthington-biochem.com/COA/
- ↑ Pitout, MJ, Nel, W. 1969. The inhibitory effect of ochratoxin a on bovine carboxypeptidase a in vitro. Biochem. Pharma. 18(8):1837-1843. DOI: 0.1016/0006-2952(69)90279-2
- ↑ Normant, E, Martres, MP, Schwartz, JC, Gros, C. 1995. Purification, cDNA cloning, functional expression, and characterization of a 26-kDa endogenous mammalian carboxypeptidase inhibitor. Proc. Natl. Acad. Sci. 92(26):12225-12229. PMCID: PMC40329
Student Contributors
- Thomas Baldwin
- Michael Melbardis
- Clay Schnell
Proteopedia Page Contributors and Editors (what is this?)
Michael Melbardis, Douglas Schnell, Thomas Baldwin, Geoffrey C. Hoops, Michal Harel