User:Francielle Aguiar Gomes/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 37: | Line 37: | ||
== Structure of Photosynthetic LH1-RC Super-complex of ''Rhodospirillum rubrum'' == | == Structure of Photosynthetic LH1-RC Super-complex of ''Rhodospirillum rubrum'' == | ||
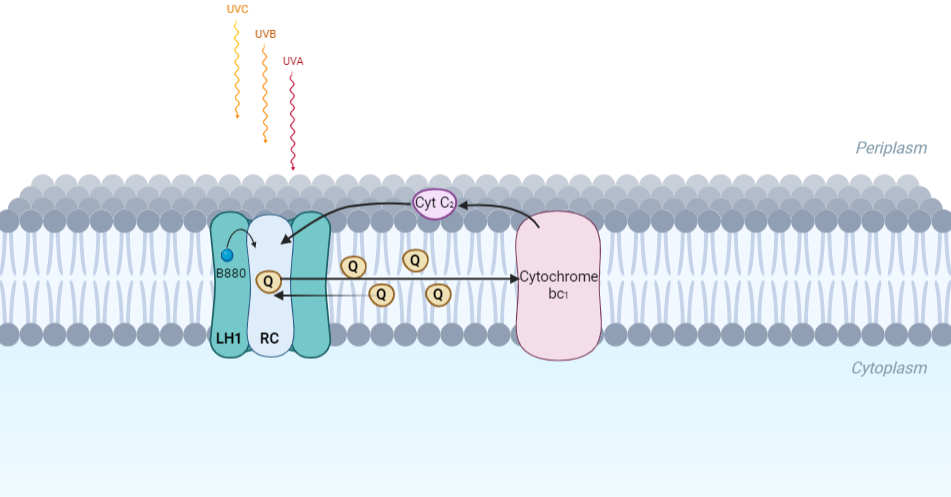
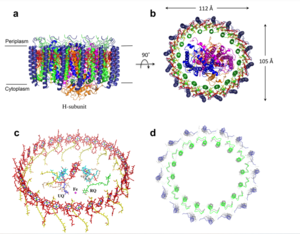
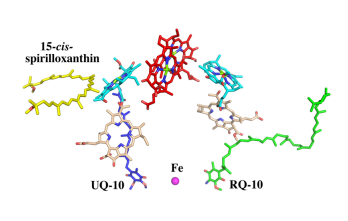
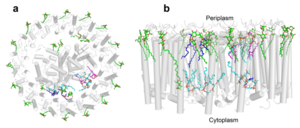
| - | The cryo-EM structure of ''Rsp. rubrum'' LH1-RC was determined at 2.76 Å resolution. The LH1 complex forms a closed, slightly elliptical double ring composed of 16 pairs of α(inner)β(outer)-polypeptides, 32 BChls aG and 16 all-trans-spirilloxanthins, as we can se on the Fig. 1. The RC of ''Rsp. rubrum'' is surrounded by the LH1 complex with only a few close contacts on the periplasmic surface with residues near Ser34 in the LH1 α-polypeptides. There is no apparent strong interactions of ''Rsp. rubrum'' RC with your LH1 polypeptides (Figure 1 a-c) as occurs in Cyt c-bound LH1-RCs where the C-terminal domains of some LH1 α-polypeptides interact extensively with the Cyt c subunit | + | The cryo-EM structure of ''Rsp. rubrum'' LH1-RC was determined at 2.76 Å resolution. The LH1 complex forms a closed, slightly elliptical double ring composed of 16 pairs of α(inner)β(outer)-polypeptides, 32 BChls aG and 16 all-trans-spirilloxanthins, as we can se on the Fig. 1. The RC of ''Rsp. rubrum'' is surrounded by the LH1 complex with only a few close contacts on the periplasmic surface with residues near Ser34 in the LH1 α-polypeptides. There is no apparent strong interactions of ''Rsp. rubrum'' RC with your LH1 polypeptides (Figure 1 a-c) as occurs in Cyt c-bound LH1-RCs where the C-terminal domains of some LH1 α-polypeptides interact extensively with the Cyt c subunit. <ref>10.1021/acs.biochem.1c00360</ref> |
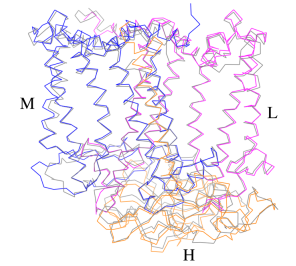
[[Image:Structure.png|300px|left|thumb| '''Fig. 1.''' Structure overview of the ''Rsp. rubrum'' LH1-RC complex. (a) Side view of the LH1-RC parallel to the membrane plane. (b) Top view of the LH1-RC from the periplasmic side of the membrane. (c) Tilted view of the cofactor arrangement. (d) Superposition of Cα carbons of the LH1 αβpolypeptides between ''Rsp. rubrum'' and <scene name='96/969634/Tch_tepidum/1'>''Tch. tepidum''</scene> (gray, PDB: 5Y5S). Color scheme: LH1-α, green; LH1-β, slate-blue; L-subunit, magenta; Msubunit, blue; BChl aG in LH1 and special pair, red sticks; Accessory BChl aG, cyan sticks; BPhe aG, light-pink sticks; Spirilloxanthin, yellow sticks; UQ10, blue sticks; RQ-10, green sticks; Fe, magenta ball. Phospholipids and detergents are omitted for clarity]] | [[Image:Structure.png|300px|left|thumb| '''Fig. 1.''' Structure overview of the ''Rsp. rubrum'' LH1-RC complex. (a) Side view of the LH1-RC parallel to the membrane plane. (b) Top view of the LH1-RC from the periplasmic side of the membrane. (c) Tilted view of the cofactor arrangement. (d) Superposition of Cα carbons of the LH1 αβpolypeptides between ''Rsp. rubrum'' and <scene name='96/969634/Tch_tepidum/1'>''Tch. tepidum''</scene> (gray, PDB: 5Y5S). Color scheme: LH1-α, green; LH1-β, slate-blue; L-subunit, magenta; Msubunit, blue; BChl aG in LH1 and special pair, red sticks; Accessory BChl aG, cyan sticks; BPhe aG, light-pink sticks; Spirilloxanthin, yellow sticks; UQ10, blue sticks; RQ-10, green sticks; Fe, magenta ball. Phospholipids and detergents are omitted for clarity]] | ||
| Line 43: | Line 43: | ||
For a more comprehensive view of the structure of ''Rsp. rubrum'' LH1-RC, this molecule can be visualized in different ways, such as, for instance, by the shape of <scene name='96/969634/Backbone/1'>backbone</scene>, <scene name='96/969634/Ballandstick/1'>Ball and Stick</scene> or <scene name='96/969634/Spacefill/1'>spacefill</scene>. | For a more comprehensive view of the structure of ''Rsp. rubrum'' LH1-RC, this molecule can be visualized in different ways, such as, for instance, by the shape of <scene name='96/969634/Backbone/1'>backbone</scene>, <scene name='96/969634/Ballandstick/1'>Ball and Stick</scene> or <scene name='96/969634/Spacefill/1'>spacefill</scene>. | ||
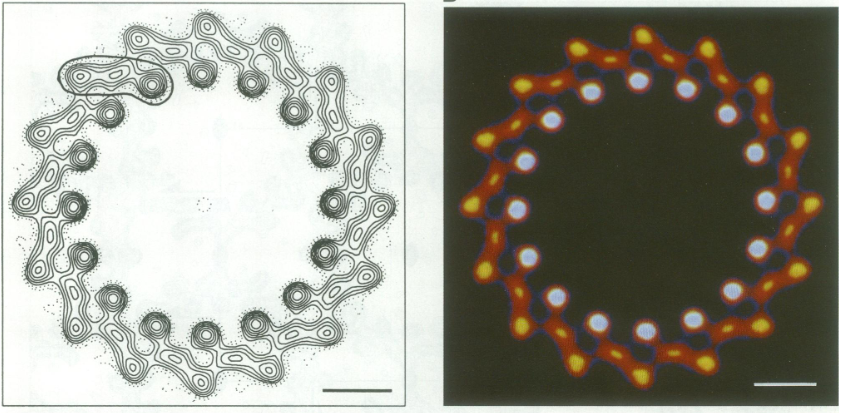
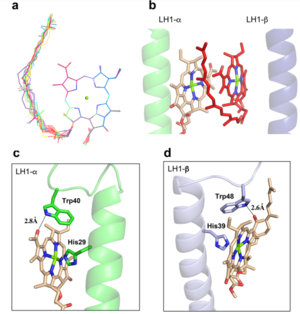
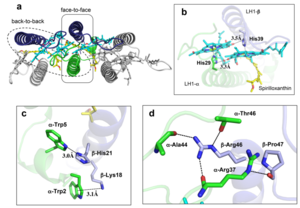
| - | The BChl aG molecules in ''Rsp. rubrum'' LH1 forms an elliptical, partially overlapping ring with average Mg−Mg distances of 9.3 Å within a dimer and 8.5 Å between dimers. These molecules are ligated by histidine residues (α-His29 and β-His39), as shown in the next image (Fig. 3). The geranylgeranyl side chains in the BChl aG associated with βpolypeptides form a tail-up conformation (Figure 2a). This may be due to the multiple double bonds present in the geranylgeranyl group, which results in a more rigid conformation. This unique conformation allows the geranylgeranyl side chains of the β-associated BChl aG to interact with the bacteriochlorin ring of the α-associated BChl aG (Figure 3b). This proximity likely allows for π−π interactions between the double bonds in the geranylgeranyl group and nearby bacteriochlorins. The C3-acetyl oxygen atoms in BChl aG form hydrogen bonds with the Trp residues (α-Trp40 and β-Trp48) of their associated polypeptides (Figure 2c, d), in agreement with the results of resonance Raman spectroscopy. The aromatic side chains of the Trp residues also interact with bacteriochlorins through π−π stacking, further stabilizing the LH1 complex. <ref>10.1073/pnas.91.15.7124</ref><ref>10.1016/0005-2728(85)90048-9</ref> | + | The BChl aG molecules in ''Rsp. rubrum'' LH1 forms an elliptical, partially overlapping ring with average Mg−Mg distances of 9.3 Å within a dimer and 8.5 Å between dimers (Figure 1c). These molecules are ligated by histidine residues (α-His29 and β-His39), as shown in the next image (Fig. 3). The geranylgeranyl side chains in the BChl aG associated with βpolypeptides form a tail-up conformation (Figure 2a). This may be due to the multiple double bonds present in the geranylgeranyl group, which results in a more rigid conformation. This unique conformation allows the geranylgeranyl side chains of the β-associated BChl aG to interact with the bacteriochlorin ring of the α-associated BChl aG (Figure 3b). This proximity likely allows for π−π interactions between the double bonds in the geranylgeranyl group and nearby bacteriochlorins. The C3-acetyl oxygen atoms in BChl aG form hydrogen bonds with the Trp residues (α-Trp40 and β-Trp48) of their associated polypeptides (Figure 2c, d), in agreement with the results of resonance Raman spectroscopy. The aromatic side chains of the Trp residues also interact with bacteriochlorins through π−π stacking, further stabilizing the LH1 complex. <ref>10.1073/pnas.91.15.7124</ref><ref>10.1016/0005-2728(85)90048-9</ref> |
[[Image:Ligant.png|300px|right|thumb| '''Fig. 2.''' (a) Superposition of the bacteriochlorin rings of 16 BChl aG molecules bound to the LH1 βpolypeptides. (b) Interactions between a geranylgeranyl side chain of the BChl aG (red sticks) bound to LH1 β-polypeptide and the bacteriochlorin ring (tint sticks) of a BChl aG bound to the LH1 α-polypeptide. (c) Coordination and hydrogen bonding of the BChl aG in LH1 α-polypeptide. (d) Coordination and hydrogen bonding of the BChl aG in LH1 β-polypeptide.]] | [[Image:Ligant.png|300px|right|thumb| '''Fig. 2.''' (a) Superposition of the bacteriochlorin rings of 16 BChl aG molecules bound to the LH1 βpolypeptides. (b) Interactions between a geranylgeranyl side chain of the BChl aG (red sticks) bound to LH1 β-polypeptide and the bacteriochlorin ring (tint sticks) of a BChl aG bound to the LH1 α-polypeptide. (c) Coordination and hydrogen bonding of the BChl aG in LH1 α-polypeptide. (d) Coordination and hydrogen bonding of the BChl aG in LH1 β-polypeptide.]] | ||
Revision as of 02:09, 26 June 2023
Photosynthetic LH1-RC Super-complex of Rhodospirillum rubrum
| |||||||||||