User:Ahmed Abdelrahman/sandbox1/ salmonella l-asparaginase
From Proteopedia
| Line 23: | Line 23: | ||
== MATERIALS AND METHODS == | == MATERIALS AND METHODS == | ||
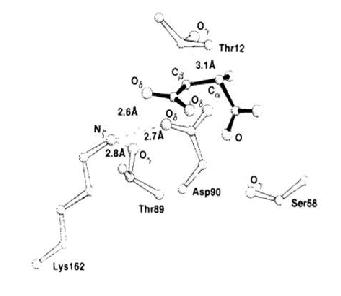
| - | Protein Blasting of Salmonella L-aspariginase II and E.coli L-asparaginase II as shown in Figure 2, was used to determine E.coli L-aspariginase II’s homology to Salmonella L-asparginase II(95%) (www.ncbi.nlm.nih.gov/blast/ 4). Figure 3 was used to determined the important amino acids that contributes in the interactions at the active site of L-asparaginase II. Rasmol version 2.6.5.1 was used to create a three-dimensional wire-frame formatted model of E.coli L-aspariginase II, once the PDB number <ref> | + | Protein Blasting of Salmonella L-aspariginase II and E.coli L-asparaginase II as shown in Figure 2, was used to determine E.coli L-aspariginase II’s homology to Salmonella L-asparginase II(95%) (www.ncbi.nlm.nih.gov/blast/ 4). Figure 3 was used to determined the important amino acids that contributes in the interactions at the active site of L-asparaginase II. Rasmol version 2.6.5.1 was used to create a three-dimensional wire-frame formatted model of E.coli L-aspariginase II, once the PDB number <ref> The RCSB Protein Data Bank, http://www.rcsb.org/pdb/explore/explore.do?structureId=3ECA, (accessed on December 1st 2009)</ref> was determined from the study done. Secondary structures such as <scene name='User:Ahmed_Abdelrahman/sandbox1/_salmonella_l-asparaginase/Alpha_helices/1'>Alpha Helices</scene> and <scene name='User:Ahmed_Abdelrahman/sandbox1/_salmonella_l-asparaginase/Beta_sheets/1'>Beta Sheets</scene> were selected in this model because they help in overall stability of the molecule due to hydrogen bonding. The amino acids illustrated are in space-fill format. The monitor lines (white) were added to increase the stability of the physical 3-Dimentional model, developed using plaster by a ZCorp 510 printer. The model shows the enzymatic activity of asparginase starts with a nucleophilic attack on the β-amide group of asparagines, leading to acyl-enzyme intermediate. Aspartate connects number of polar side chains to anchor with the product. The main-chain nitrogen and the Oɣ of <scene name='User:Ahmed_Abdelrahman/sandbox1/_salmonella_l-asparaginase/Thr89/2'>Thr89</scene> form hydrogen bonds with the β-carboxyl group of aspartate. <scene name='User:Dominic_Fiorentino/Sandbox1_Salmonella_L-asparaginase_II/Thr89/1'>Thr89</scene> is the most likely candidate as the nucleophile that attacks the aspartate. The only basic residue in the active site region that could influence the nucleophilic character of <scene name='User:Ahmed_Abdelrahman/sandbox1/_salmonella_l-asparaginase/Thr89/3'>Thr89</scene> is <scene name='User:Ahmed_Abdelrahman/sandbox1/_salmonella_l-asparaginase/Lys162/1'>Lys162</scene>. The positions of <scene name='User:Dominic_Fiorentino/Sandbox1_Salmonella_L-asparaginase_II/Thr12/1'>Thr12</scene> stabilize the tetrahedral intermediate formed between <scene name='User:Ahmed_Abdelrahman/sandbox1/_salmonella_l-asparaginase/Thr89/3'>Thr89</scene> and its substrates (aspartate). The nitrogen atom of aspartate forms hydrogen bonds with other amino acids, from the adjacent intimately bound subunit. These hydrogen bonds facilitate the binding of the substrate and stabilization of the transition states. <scene name='User:Ahmed_Abdelrahman/sandbox1/_salmonella_l-asparaginase/Ser58/1'>Ser58</scene> was chosen within the model because the oxygen of serine has interactions with the hydrogen of the alpha-carboxyl group. Aspartic acid (<scene name='User:Ahmed_Abdelrahman/sandbox1/_salmonella_l-asparaginase/Asp90/1'>Asp90</scene>) was highlighted in red, in the model because it influences the basicity of <scene name='User:Ahmed_Abdelrahman/sandbox1/_salmonella_l-asparaginase/Lys162/1'>Lys162</scene> amino acid which directly affects the nucleophilic character of <scene name='User:Ahmed_Abdelrahman/sandbox1/_salmonella_l-asparaginase/Thr89/3'>Thr89</scene>. These interactions were chosen to be demonstrated because the Serine acts as a nucleophile in this enzymatic reaction. However, the Serine will only act as a proper nucleophile if the aspartate is opposite orientation to the asparagene (enzyme) insinuating a reaction by <scene name='User:Ahmed_Abdelrahman/sandbox1/_salmonella_l-asparaginase/Ser58/1'>Ser58</scene> nucleophilically binding to the alpha-carboxyl group <ref name="swain"/> |
Revision as of 21:46, 9 December 2009
Inhibition of TCR-β surface expression by L-asparaginase II found in S. Typhimurium
PDB#3ECA
Contents |
ABSTRACT
Salmonella enterica serovar Typhimurium (S.Typhimurium) uses L-asparaginase II to evade the development of T cell-mediated adaptive immunity by interfering with T cell priming; the activation, proliferation, and differentiation of naïve T cells[1]. The complete structure of L-asparaginase II of salmonella is not fully determined, although it is 95% homologous to L-asparaginase of Escherichia coli, which is determined by protein blasting. In such an alignment, amino acids at positions important for a particular function are expected to be well conserved within, but different between, the functional classes. The amino acid alignment between the two homologous proteins suggests a similarity of their mechanisms of action. In addition, a comparative strategy can identify the specificity determining positions of the two enzymes. The study identifies the active site of L-asparaginase of E.coli and positions near it that affects its function. E.coli L-asparaginase II is a tetramer with two identical active domains A and C [2]. L-asparaginase has been used for treatment of acute lymphoblastic leukemia. The structural model created using RasMol displays the active site of chain A with specific colors to indicate secondary structures such as beta sheets and alpha helices and specific amino acids. The hydrogen bonds of the alpha helices and beta sheets help in the overall stability of molecule. Specific amino acids such as , , , , and , that are displayed in the model interact in the active site. The experimental study also confirmed that addition of E.coli L-asparaginase II to an uninfected subject down-modulates TCR-B expression and the enzyme L-asparaginase II is used clinically for the treatment of acute lymphoblastic leukemia acting as a possible tumor suppressor. It is possible that the enzyme L-asparaginase II plays a vital role in the delayed immune response of T cells when encountered by Salmonella. Future experiments can be performed to study inhibition of Salmonella L-asparginase II since its structure is not available in PDB form, the use of E.coli L-Asparginase II can be used instead due to homology. Inhibition of L-asparaginase II returns the infected subject to the normal rate of immune response and creates higher expression TCR-B receptors on the surface when compared to a wild/type infected subject; this in turn, creates more T cells and a normal immune response.
|
INTRODUCTION
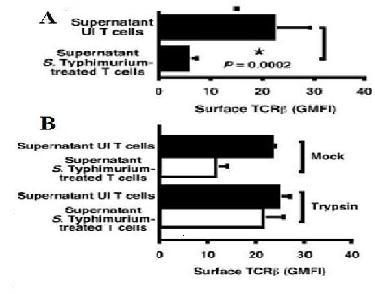
T cell mediated adaptive immunity is required to fight against infections with an intracellular facultative bacterial pathogen, Salmonella enterica serovar Typhimurium (S. Typhimurium). Macrophages and neutrophils are inflammatory cells of the immune system that provide the first line of defense against pathogenic microorganisms. Lymphocytes such as T cells are the ultimate response to clear the microbial infection. The study shows that the immune response to the S. Typhimurium is slow and in-efficient which reduces the amount of surface and intracellular TCR-B proteins as seen by the down modulation of TCR-B protein [a receptor molecule required for Ag (silver) recognition and T cell function] when cultured in presence of S.Typhimurium. The contact between S. Typhimurium and T cells reduces the expression of TCR-B that is secreted. Furthermore, the study explores why S. Typhimurium plays an active role in inhibiting development of protective immunity. A genetic screen was used to identify the salmonella genes required for T cell inhibition. This screen identified the gene ansB which codes for L-asparaginase II [1]. L-asparaginase II produced by the Salmonella, catalyzes the hydrolysis of L-asparagine to L-aspartate and ammonia. Type I asparaginase is located in the cytosol, whereas type II asparaginase in located in the peri-plasmic region (the space between the inner and outer membrane) of the bacteria and has a much higher affinity for asparagines. Thus, the RasMol program is used to create a model of wild type E.coli L asparaginase II enzyme showing and , and amino acids such as , , , , and to study the structure of L-asparaginase II of salmonella . Since the structure of Salmonella L-asparaginase II is not determined which is 95% homologous (determined by blasting) to E.coli L-asparaginase II since the structure of L-asparaginase II of salmonella is still not developed (www.ncbi.nlm.nih.gov/blast/5) If an uninfected subject is administered with E.coli L-asparaginase II, it causes down modulation of TCR-B expression as seen in the wild type S. Typhimurium infected subject.
Figure 1[1] was determined from the study done. Secondary structures such as and were selected in this model because they help in overall stability of the molecule due to hydrogen bonding. The amino acids illustrated are in space-fill format. The monitor lines (white) were added to increase the stability of the physical 3-Dimentional model, developed using plaster by a ZCorp 510 printer. The model shows the enzymatic activity of asparginase starts with a nucleophilic attack on the β-amide group of asparagines, leading to acyl-enzyme intermediate. Aspartate connects number of polar side chains to anchor with the product. The main-chain nitrogen and the Oɣ of form hydrogen bonds with the β-carboxyl group of aspartate. is the most likely candidate as the nucleophile that attacks the aspartate. The only basic residue in the active site region that could influence the nucleophilic character of is . The positions of stabilize the tetrahedral intermediate formed between and its substrates (aspartate). The nitrogen atom of aspartate forms hydrogen bonds with other amino acids, from the adjacent intimately bound subunit. These hydrogen bonds facilitate the binding of the substrate and stabilization of the transition states. was chosen within the model because the oxygen of serine has interactions with the hydrogen of the alpha-carboxyl group. Aspartic acid () was highlighted in red, in the model because it influences the basicity of amino acid which directly affects the nucleophilic character of . These interactions were chosen to be demonstrated because the Serine acts as a nucleophile in this enzymatic reaction. However, the Serine will only act as a proper nucleophile if the aspartate is opposite orientation to the asparagene (enzyme) insinuating a reaction by nucleophilically binding to the alpha-carboxyl group [2]
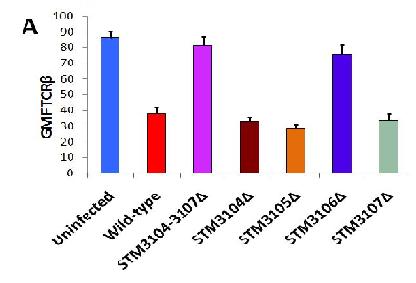
Figure 5[2]: A. Reveals how modification of STM 3104-3107 and just STM-3106, both have an identical effect in TCR-B expression. This shows that L-asparaginase II (STM 3106) is required for down-modulation of TCR-B expression
B. Indicates Salmonella Typhimurium (STM) gene coding associated with the actual name of the gene.
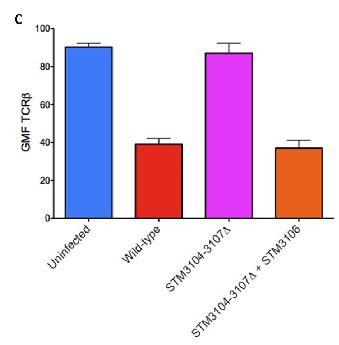
C. Demonstrates that if STM-3104,3105, 3107 is deleted along with a normal functioning stm-3106, it will still down-modulate TCR-B expression.
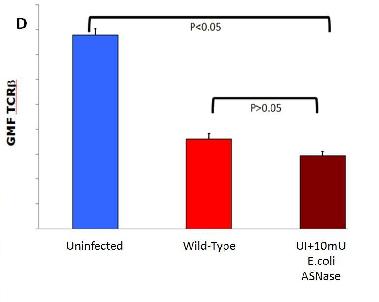
D. Shows how addition of E.coli L-asparaginase II to an uninfected subject will down-modulate TCR-B expression.
Figure 3 [2] Structure that was used to determine the important amino acids to be highlighted in the RasMol model of E.coli L-asparaginase II. It demonstrates the interaction of the Asparate substrate (solid bonds) with the neighboring amino acids such as Thr 89, Thr12 and Ser58. In a chain of reactions, Asp90 affects Lys162 which influences the nucleophilicity and basicity of Thr89.
Figure 2[3] Protein Blast of Salmonella L-aspariginase II and E.coli L-asparaginase II, revealing 95% homology.
DISCUSSION AND CONCLUSION
The immune system is an essential component of the human body which serves to protect itself from foreign pathogens and substances. The widely known bacterial pathogen called Salmonella has been shown to induce a delayed immune response in T cells. Hence, it acts as a direct effecter for TCR-B down modulation. After genetic screening revealed that the gene required to down modulate TCR B expression is L-asparaginase II, reasoning for its function was further studied [1],[4]. The L-asparaginase II enzyme has an anti-leukemic function and is used clinically for the treatment of acute lymphoblastic leukemia. Hence, by acting as a possible tumor suppressor, it is possible that it plays a role in the delayed immune response of T cells when encountered by Salmonella [2]. With the entire structure of Salmonella L-asparaginase II not yet developed, the structurally and functionally similar tetramer of L-asparaginase II of E.coli (95% homologous to Salmonella L-asparaginase II) was used to convey how this enzyme could act correspondingly as a suppressor of the immune system. Possible future experiments can be done to test how to inhibit Salmonella L-asparginase II in the E.coli molecule since there are no structures of Salmonella L-asparginase II available in PDB form. Inhibition of L-asparaginase II returns to the normal rate of immune response and creates higher expression TCR-B receptors on the surface when compared to a wild/type infected subject; this in turn, creates more T cells and a normal immune response.
REFERENCES
- ↑ 1.0 1.1 1.2 1.3 van der Velden AW, Dougherty JT, Starnbach MN. Down-modulation of TCR expression by Salmonella enterica serovar Typhimurium. J Immunol. 2008 Apr 15;180(8):5569-74. PMID:18390741
- ↑ 2.0 2.1 2.2 2.3 2.4 Swain AL, Jaskolski M, Housset D, Rao JK, Wlodawer A. Crystal structure of Escherichia coli L-asparaginase, an enzyme used in cancer therapy. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1474-8. PMID:8434007
- ↑ national institute of health, http://www.ncbi.nlm.nih.gov/blast/, (accessed on December 1st 2009)
- ↑ Forbes SJ, Eschmann M, Mantis NJ. Inhibition of Salmonella enterica serovar typhimurium motility and entry into epithelial cells by a protective antilipopolysaccharide monoclonal immunoglobulin A antibody. Infect Immun. 2008 Sep;76(9):4137-44. Epub 2008 Jul 14. PMID:18625740 doi:10.1128/IAI.00416-08