Odorant binding protein
From Proteopedia
| Line 2: | Line 2: | ||
Odorant-binding protein (OBP) are soluble proteins which involve in the processes of odorant detection in the olfactory sensilla. | Odorant-binding protein (OBP) are soluble proteins which involve in the processes of odorant detection in the olfactory sensilla. | ||
| - | + | Though functionally same, vertebrates and insects OBP have different origin and structure. | |
| - | Though | + | |
OBPs are important for insect olfaction. For instance, OBP76a (LUSH) in the fly [http://en.wikipedia.org/wiki/Drosophila_melanogaster ''Drosophila melanogaster''] is required for the detection of the pheromone vaccenyl acetate [Ha and Smith, 2006; Xu et al., 2005] and has been proven to adopt a conformation that activates the odorant receptor [Laughlin et al., 2008]. | OBPs are important for insect olfaction. For instance, OBP76a (LUSH) in the fly [http://en.wikipedia.org/wiki/Drosophila_melanogaster ''Drosophila melanogaster''] is required for the detection of the pheromone vaccenyl acetate [Ha and Smith, 2006; Xu et al., 2005] and has been proven to adopt a conformation that activates the odorant receptor [Laughlin et al., 2008]. | ||
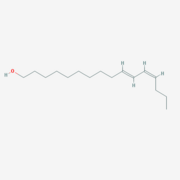
[[Image:Bombykol.png|thumb|upright=1|Bombykol, a sex pheromone of ''Bombyx mori'', from [http://pubchem.ncbi.nlm.nih.gov/compound/Bombykol#section=Top PubChem]]] | [[Image:Bombykol.png|thumb|upright=1|Bombykol, a sex pheromone of ''Bombyx mori'', from [http://pubchem.ncbi.nlm.nih.gov/compound/Bombykol#section=Top PubChem]]] | ||
| Line 31: | Line 30: | ||
<StructureSection load='1ls8' size='340' side='right' caption='''Bombyx mori'' PBP -BmorPBP scene=''> | <StructureSection load='1ls8' size='340' side='right' caption='''Bombyx mori'' PBP -BmorPBP scene=''> | ||
Pheromone binding proteins (PBPs) are specialized members of the insect odorant-binding protein (OBP) super-family. | Pheromone binding proteins (PBPs) are specialized members of the insect odorant-binding protein (OBP) super-family. | ||
| - | The main | + | The main purpose in the dult moth's short life is reproduction. In fact, the male and female moth invest all of theire energy and resourses hoping to reach to the ultimate goal- mating. This long journey begins when the female moth releases a sex pheromone, usualy in specific hours in the night <ref>doi: 10.1007/BF01946910</ref>. |
| - | BmorPBP was first identified in the ''B. mori'' male antennae by Krieger et al. in 1996 <ref>doi: 10.1016/0965-1748(95)00096-8</ref>, as the PBP of the first sex pheromone discovered ((E,Z)-10,12-hexadecadienol, or [[http://en.wikipedia.org/wiki/Bombykol Bombykol]]). The male moth needs to detect | + | BmorPBP was first identified in the ''B. mori'' male antennae by Krieger et al. in 1996 <ref>doi: 10.1016/0965-1748(95)00096-8</ref>, as the PBP of the first sex pheromone discovered ((E,Z)-10,12-hexadecadienol, or [[http://en.wikipedia.org/wiki/Bombykol Bombykol]]). The male moth needs to detect minute amount of the pheromone in the air, while following turbulent wind-born pheromone trail and response fast (experimental evidence shows a response time of 0.5 seconds<ref>doi: 10.1038/293161a0</ref>). As the electrical signal transduction after the activation of the odorant receptor are too fast, Kaissling <ref name="Kaissling 2009">doi: 10.1007/s00359-009-0461-4</ref>have suggested that the |
====BmorPBP structure and function==== | ====BmorPBP structure and function==== | ||
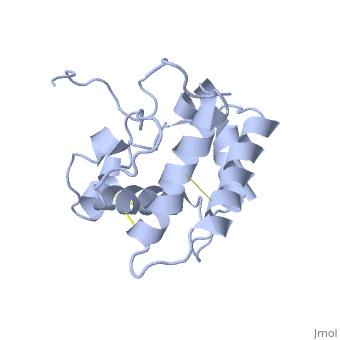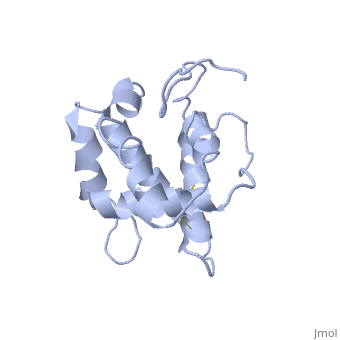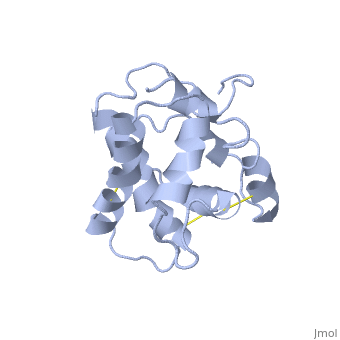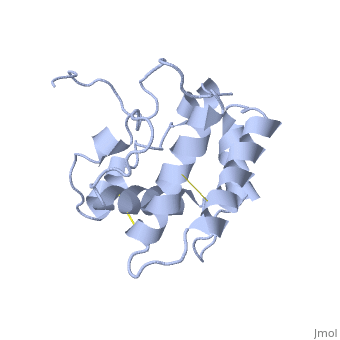
| - | The protein has 164 amino acids that forms 6-7 alpha helices. Three disulfide bonds (in yellow) formed by <scene name='68/683383/Cysteins6/1'>6 cystein </scene> residues tied four helices, and form the compact and robust | + | The protein has 164 amino acids that forms 6-7 alpha helices. Three disulfide bonds (in yellow) formed by <scene name='68/683383/Cysteins6/1'>6 cystein </scene> residues tied four helices, and form the compact and robust structure of the protein. As expected from a soluble protein, its surface is covered with <scene name='68/683383/Charged_residues/1'>charged residues</scene>, which allows it to make interactions with the water molecule and solubilize in the sensillar lymph. |
====BmorPBP ligand and ligand binding==== | ====BmorPBP ligand and ligand binding==== | ||
| - | The protein natural ligand is the moth pheromone <scene name='68/683383/Bombykol_ligand_in_2p71/1'>Bombykol</scene>. However, it was demonstrated that other molecules can also bound to the protein cavity <ref>doi: 10.1016/j.str.2007.07.013</ref>. The interaction with the ligand is | + | The protein natural ligand is the moth pheromone <scene name='68/683383/Bombykol_ligand_in_2p71/1'>Bombykol</scene>. However, it was demonstrated that other molecules can also bound to the protein cavity <ref>doi: 10.1016/j.str.2007.07.013</ref>. The interaction with the ligand is being made by 4 alpha helices 1, 4, 5 and 6 in the core of the protein, which form the binding cavity <ref>doi: 10.1016/S1074-5521(00)00078-8</ref>. |
Inside the binding cavity, <scene name='68/683383/Residues_interacting/1'>non-charged residues</scene> are interacting with the pheromone, mainly by van der waals bounds. Out of those residues, some are conserved across OBP of lepidopteran (<font color='green'><b>in green</b></font>), and the rest are conserved in lepidopteran PBP only (<font color='light blue'><b>in light blue</b></font>). | Inside the binding cavity, <scene name='68/683383/Residues_interacting/1'>non-charged residues</scene> are interacting with the pheromone, mainly by van der waals bounds. Out of those residues, some are conserved across OBP of lepidopteran (<font color='green'><b>in green</b></font>), and the rest are conserved in lepidopteran PBP only (<font color='light blue'><b>in light blue</b></font>). | ||
In addition, the hydroxyl group of the pheromone bombykol forms a <scene name='68/683383/Ser56_interaction_with_oxg/2'>hydrogen bond with the sidechain of Ser56</scene>, Ser56 in red, oxygens are in purple (O–O distance of 2.8 Å). | In addition, the hydroxyl group of the pheromone bombykol forms a <scene name='68/683383/Ser56_interaction_with_oxg/2'>hydrogen bond with the sidechain of Ser56</scene>, Ser56 in red, oxygens are in purple (O–O distance of 2.8 Å). | ||
| Line 44: | Line 43: | ||
====Protein conformations==== | ====Protein conformations==== | ||
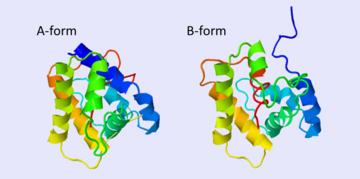
[[Image:A -B forms without ligand.png|thumb|upright=2|The A and B forms without ligand (PDB IDs: [[1gm0]] and [[1ls8]]).]] | [[Image:A -B forms without ligand.png|thumb|upright=2|The A and B forms without ligand (PDB IDs: [[1gm0]] and [[1ls8]]).]] | ||
| - | BmorPBP has two conformations: The "open form" (A) and the "close form" (B)<ref>DOI: 10.1074/jbc.274.43.30950</ref>. The bombykol and the alpha-helix loacated in the c-terminus of the protein compete for the binding site: when the c-terminus is inside the binding cavity it get's an alpha helix shape, and the protien is in its "close form" (B), | + | BmorPBP has two conformations: The "open form" (A) and the "close form" (B)<ref>DOI: 10.1074/jbc.274.43.30950</ref>. The bombykol and the alpha-helix loacated in the c-terminus of the protein compete for the binding site: when the c-terminus is inside the binding cavity it get's an alpha helix shape, and the protien is in its "close form" (B), whereas in the "open form" (A) the c-terminus is outside of the protein and has no defined secondary structure. Binding experiments have shown that the B-form binds 15 times higher than the A-form <ref>doi: 10.1073/pnas.0501447102</ref>, therefore considered to be the carrier of the pheromone. The complex of the A-form and the pheromone, is then considered the form that activates the receptor. |
| - | The <scene name='68/683383/1dqe-1gm0/2'>transition between the two | + | The <scene name='68/683383/1dqe-1gm0/2'>transition between the two conformation</scene> is both pH and ligand dependent <ref>doi: 10.1073?pnas.251532998</ref><ref>DOI: 10.1016/j.bbrc.2005.07.176</ref><ref>doi: 10.1073/pnas.1317706110</ref>. In short, the B-form (c-terminus outside the cavity) occurs only at neutral pH and in the presence of the ligand. The A-form (c-terminus inside the cavity) occurs at both low and neutral pH, yet at the latter only in the absence of ligand. Therefore, in neutral pH when the ligand is binding to the protein in its A-form, the complex formation causes a change in conformation to the B-form. However, both A and B forms are equally distributed in the lymph. |
{{Button Toggle AnimationOnPause}} | {{Button Toggle AnimationOnPause}} | ||
'''Conformation transition mechanism:''' | '''Conformation transition mechanism:''' | ||
| - | The c-terminus of the protein bears mostly | + | The c-terminus of the protein bears mostly non-polar amino acids. Yet on the surface of the helix there are three exceptional amino acids: Asp-132, Glu-137, and Glu-141, which are conserved in moth PBP <ref>doi: 10.1016/j.bbrc.2005.07.176</ref>. Of these, residues <scene name='68/683383/Bombykol_ligand_in_2p71/2'>Asp-132</scene> (and Glu-141, if present) triggers the formation of the alpha-helix upon protonation at low pH. This causes the ejaculation of the ligand from the binding pocket, which is replaced by the formatted alpha helix<ref>doi: 10.1016/j.bbrc</ref>. |
| - | Studies on other Lepidopterans that show a | + | Studies on other Lepidopterans that show a similar pH dependent conformation suggests that this model is a general model moth PBP<ref name="Leal" />. |
| - | Nonetheless, the | + | Nonetheless, the enormous diversity among insects is not allowing us to assume this model is true for all insects' OBPs. |
====Receptor activation==== | ====Receptor activation==== | ||
Two theories have been propsed for the activation of the odorant receptors located on the dendrtirte membrane. One theory suggests that the pheromone-PBP complex is needed for the receptor activation, while the second theory argue that the pheromone itself is sufficient for the activation of the receptor. | Two theories have been propsed for the activation of the odorant receptors located on the dendrtirte membrane. One theory suggests that the pheromone-PBP complex is needed for the receptor activation, while the second theory argue that the pheromone itself is sufficient for the activation of the receptor. | ||
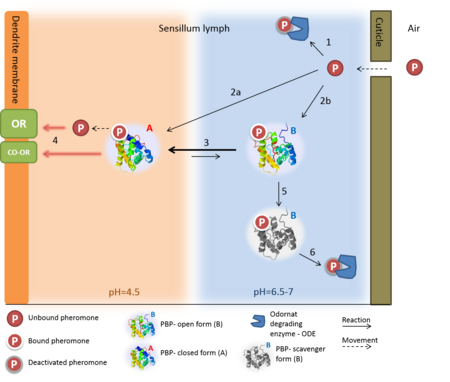
| - | [[Image:N model extended.png|thumb|upright=2.5|Figure 1. The events prior the neuron excitation, following the "N model" suggested by Kaissling (2009)<ref name="Kaissling 2009" />: The pheromone enters the sensillar lymph through a pore in cuticle. Following | + | [[Image:N model extended.png|thumb|upright=2.5|Figure 1. The events prior the neuron excitation, following the "N model" suggested by Kaissling (2009)<ref name="Kaissling 2009" />: The pheromone enters the sensillar lymph through a pore in cuticle. Following entrance the pheromone can be degraded by the ODE (1) -or- bind to the A and B protein forms (2a and 2b, respectively). The transition between the two forms in the neutral pH of the lymph, is in favor for the B form in the presence of the ligand (3). However, when the complex arrives at the low pH near the membrane, the transition is in favor of the A-form, (4) in which the pheromone is ejaculated due to the change in conformation- c-terminus is pushing out the ligand. The activation of the complex of odorant receptor and coreceptor (OR:OR-CO), is induced by ether the complex of pheromone-PBP, or by the pheromone alone (5, two options). The B-form can also act as a scavenger, as it mediates the deactivation of the pheromone (6) and releases it to the ODE (7)]] |
*'''Activation by the pheromone alone''' | *'''Activation by the pheromone alone''' | ||
| - | This model is supported by the pH dependent | + | This model is supported by the pH dependent conformation transition, that is described above. The bulk of the sensillar lymph is in neutral pH (6.5-7), while environment near the dendrite membrane bears a low pH (4.5), due to the negative charges on the surface of the membrane <ref>DOI: 10.1016/0040-8166(84)90004-1</ref>, which cause the accumulation of positively charged kations near the membrane surface (20-50 nm)<ref name="Kaissling 2009" />. According to this model (illustrated in [[figure 1]]), the pheromone is entering the sensillar lymph through a pore in the cuticle, then it can be either degraded by odorant degrading enzymes (ODE) or bind to a PBP (of both forms). Once the complex is arriving to the low pH environment near the dendrite membrane the PBP will shift to the A-form, thereby ejaculating the ligand from the binding pocket, allowing it to activate the OR:CO-OR complex and the cellular signal transduction begins. |
| Line 71: | Line 70: | ||
== See also == | == See also == | ||
*[[Odorant_binding_protein_3D_structures]] | *[[Odorant_binding_protein_3D_structures]] | ||
| - | *[[Chemical | + | *[[Chemical communication in arthropods]] |
*[[Pheromone binding protein]] | *[[Pheromone binding protein]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 14:04, 14 January 2015
Contents |
Introduction
Odorant-binding protein (OBP) are soluble proteins which involve in the processes of odorant detection in the olfactory sensilla.
Though functionally same, vertebrates and insects OBP have different origin and structure. OBPs are important for insect olfaction. For instance, OBP76a (LUSH) in the fly Drosophila melanogaster is required for the detection of the pheromone vaccenyl acetate [Ha and Smith, 2006; Xu et al., 2005] and has been proven to adopt a conformation that activates the odorant receptor [Laughlin et al., 2008].
OBP in insects
OBP Function
Despite five decades of intensive research, the exact roles of OBP and the mechanism by which the odorant receptor (OR) is activated are still in dispute [1][2].
A few functions have been suggested for OBP: 1. Solubelizing the odorant molecule and its transportation in the sensillar lymph.
2. Protecting the odorant molecule from the odorant degrading enzymes, in the sensillar lymph.
3. Activating of the odorant receptor on the dendrite membrane, by the odorant-OBP complex.
4. Mediating the deactivation of the odorant molecule after the activation of the receptor.
5. An organic anion (the protein has 9 negative charges).
Of all, the first role of OBP as an odorant solubilizer and carrier is generally accepted.
In order to explain the structure and function of these fascinating proteins, this page will further focus on a particular OBP - the well investigated Bombyx mori PBP: BmorPBP.
Bombyx mori BmorPBP (lets talk about sex..)
| |||||||||||
See also
- Odorant_binding_protein_3D_structures
- Chemical communication in arthropods
- Pheromone binding protein
References
Proteopedia Page Contributors and Editors (what is this?)
Nurit Eliash, Michal Harel, Joel L. Sussman, Alexander Berchansky, Jaime Prilusky