User:Kurt Corsbie/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
===Biological Background=== | ===Biological Background=== | ||
G protein-coupled receptors (GPCRs) can be broken up into four classes: A, B, C, and F. <ref name="GPCR">PMID:23407534</ref>. Metabotropic glutamate receptor 5 (mGLu<sub>5</sub>) is a Class C GPCR that couples with the G<sub>q</sub> pathway <ref name="Pinj">PMID:12782243</ref>. | G protein-coupled receptors (GPCRs) can be broken up into four classes: A, B, C, and F. <ref name="GPCR">PMID:23407534</ref>. Metabotropic glutamate receptor 5 (mGLu<sub>5</sub>) is a Class C GPCR that couples with the G<sub>q</sub> pathway <ref name="Pinj">PMID:12782243</ref>. | ||
| - | ===Importance=== | ||
| - | == Function == | ||
== Structure == | == Structure == | ||
| + | === Overall Stucture === | ||
| + | mGlu<sub>5</sub> is seen as a [https://en.wikipedia.org/wiki/Protein_dimer homodimer] ''in vivo,'' with each subunit comprising three domains; extracellular, trans-membrane and cysteine-rich. The structures shown here are centered on the trans-membrane domain, comprised of seven α-helices all roughly parallel to one another <ref name="Primary">PMID: 25042998 </ref>. Also displayed are Intracellular Loop (ICL) 1 forming a short α-helix, ICL3 and Extracellular Loops (ECL) 1 and 3, which lack secondary structure, and ECL2, which interacts with trans-membrane (TM) helices 1,2 and 3 and ECL 1<ref name="Primary">PMID: 25042998 </ref>. | ||
| + | ===Key Interactions=== | ||
| + | A number of intramolecular interactions within the trans-membrane domain stabilize the inactive conformation of mGlu<sub>5</sub>. The first of these interactions is an ionic interaction, termed the <scene name='72/726409/Ionic_lock2/1'>Ionic Lock </scene>, between Lysine 665 of TM3 and Glutamate 770 of TM6. Evidence for the importance of this interaction came through a kinetic study of mutant proteins where both residues were separately mutated to alanine resulting in constitutive activity of the GPCR and its coupled pathway<ref name="Primary">PMID: 25042998 </ref>.A second critical interaction that stabilizes the inactive conformer is a <scene name='72/726409/Hydrogen_bond_614-668/1'>Hydrogen Bond </scene> between Serine 614 of ICL1 and Arginine 668 of TM3. Similarly, when Serine 614 is mutated to alanine high levels of activity are seen in the mutant GPCR<ref name="Primary">PMID: 25042998 </ref>. | ||
| + | A <scene name='72/726404/Scene_6/3'>Disulfide Bond </scene> between Cystine 644 of TM3 and Cystine 733 of ECL2 is critical at anchoring ECL2 and is highly conserved across Class C GPCR’s<ref name="Primary">PMID: 25042998 </ref>. The ECL2 position, in combination with the helical bundle of the trans-membrane domain, create a <scene name='72/726409/Electrogradient2/3'>Binding Cap</scene> that restricts entrance to the allosteric binding site within the seven trans-membrane α-helices. This restricted entrance has no effect on the natural ligand, glutamate, as it binds to the extracellular domain, but dictates potential drug targets that act through allosteric modulation<ref name="Primary">PMID: 25042998 </ref>. | ||
| + | ===Comparison with Class A and B GPCRs=== | ||
| + | The structure of the trans-membrane domain of mGlu<sub>5</sub> was compared to rhodopsin, a class A GPCR, and CRF<sub>1</sub>R, a class B GPCR. The superimposition of all three structures demonstrated the greatest divergence occurs across the top half of the trans-membrane bundle, owning to the different extracellular domains that accompany each GPCR class. Furthermore, mGlu<sub>5</sub> TM7 is shifted inwards 5 Angstrom compared to the class A GPCR and TM5 is shifted inwards 6 Angstrom compared to both GPCRs, narrowing the allosteric binding entrance to a greater extent <ref name="Primary">PMID: 25042998 </ref>. | ||
| + | == Clinical Relevance == | ||
| + | ===Role in Diseases=== | ||
| + | mGlu<sub>5</sub> is located mainly post-synaptically and is in high abundance in the [https://en.wikipedia.org/wiki/Nucleus_accumbens nucleus accumbens], [https://en.wikipedia.org/wiki/Caudate_nucleus caudate nucleus], [https://en.wikipedia.org/wiki/Striatum striatum], [https://en.wikipedia.org/wiki/Hippocampus hippocampus] and [https://en.wikipedia.org/wiki/Cerebellum cerebellar cortex] <ref name="Local">PMID: 8295733 </ref>.These areas of the brain are highly involved in cognition, motivation and emotion, essential neural functions for everyday life. Diseases and other mental deficiencies arise from either an over activation of the GPCR, which over activates its coupled signaling pathway, or from under activation of both. Negative allosteric modulators (NAMs) work to decrease protein activity and are being studied as treatments for [https://en.wikipedia.org/wiki/Fragile_X_syndrome fragile X-syndrome], depression, anxiety and [https://en.wikipedia.org/wiki/Dyskinesia dyskinesia]. Conversely Positive allosteric modulators work to increase protein activity and are being studied for the treatment of schizophrenia and cognitive disorders<ref name="Diseases">PMID: 24237242</ref>. | ||
| + | |||
| + | |||
| + | ===Interactions with a Negative Allosteric Modulator=== | ||
| + | The structure of mGlu<sub>5</sub> bound to the NAM [https://en.wikipedia.org/wiki/Mavoglurant Mavoglurant] demonstrates how protein activity is decreased through drug interactions. | ||
| + | <scene name='72/726404/Scene_7/2'>Mavoglurant</scene> binds within the core of the seven trans-membrane α-helices, forming multiple interactions with the protein that further stabilize the inactive conformation. | ||
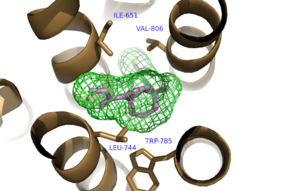
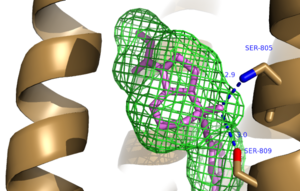
| + | The bicyclic ring system of the drug is surrounded by a pocket of mainly hydrophobic residues including Val 806, Met 802, Phe 788, Trp 785, Leu 744, Ile 651, Pro 655 and Asn 747<ref name="Primary">PMID: 25042998 </ref> (Figure 1). The carbamate tail of mavoglurant forms a hydrogen bond through its carbonyl oxygen to the amide side-chain of Asparagine 747 of TM4 (Figure 2). A hydroxyl group similarly forms hydrogen bonds to the protein, specifically to two serine residues (S805 and S809) of TM7 which form a hydrogen bonding network to other residues through their main chain atoms and a coordinated water molecule (omitted for clarity) (Figure 3). The interactions between Mavoglurant andmGlu<sub>5</sub> involved TM helices that were not previously stabilized by any strong interactions, introducing a new level of stability that favors the inactive conformation of the protein and hence decrease activity<ref name="Primary">PMID: 25042998 </ref>. | ||
| + | [[Image:Mav_Hydrophobic_pocket.png |300 px|left|thumb|Figure 1.Hydrophobic Pocket Surrounding Mavoglurant]] | ||
| + | [[Image:Mav_HB_1.1.png|300 px|left|thumb|Figure 2.Hydrogen Bonding interactions between protein and Mavoglurant]] | ||
| + | [[Image:Mav_HB_2.png|300 px|left|thumb|Figure 3. Further Hydrogen Bonding between protein and Mavoglurant]] | ||
| + | <scene name='72/726409/Mavoglurant_overview/1'>Mavoglurant Stabilzed by ECL2</scene> | ||
| + | </StructureSection> | ||
[[Image:Mav_Hydrophobic_pocket.png |300 px|left|thumb|Hydrophobic Pocket Surrounding Mavoglurant]] | [[Image:Mav_Hydrophobic_pocket.png |300 px|left|thumb|Hydrophobic Pocket Surrounding Mavoglurant]] | ||
| Line 29: | Line 48: | ||
<scene name='72/726409/Mavoglurant_overview/1'>Mavoglurant Stabilized by ECL2</scene> | <scene name='72/726409/Mavoglurant_overview/1'>Mavoglurant Stabilized by ECL2</scene> | ||
| - | |||
| - | == Disease == | ||
| - | |||
| - | == Relevance == | ||
| - | |||
| - | == Structural highlights == | ||
| - | |||
Revision as of 17:32, 28 March 2016
| |||||||||||
Image:Mav HB 1.1.png
Hydrogen Bonding interactions between protein and Mavoglurant
Domain highlights
</StructureSection>
References
- ↑ Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, Brown A, Rodriguez SS, Weller JR, Wright AC, Bergmann JE, Gaitanaris GA. The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci U S A. 2003 Apr 15;100(8):4903-8. Epub 2003 Apr 4. PMID:12679517 doi:http://dx.doi.org/10.1073/pnas.0230374100
- ↑ 2.0 2.1 Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013 Feb 14;494(7436):185-94. doi: 10.1038/nature11896. PMID:23407534 doi:http://dx.doi.org/10.1038/nature11896
- ↑ Pin JP, Galvez T, Prezeau L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol Ther. 2003 Jun;98(3):325-54. PMID:12782243
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 4.8 4.9 Dore AS, Okrasa K, Patel JC, Serrano-Vega M, Bennett K, Cooke RM, Errey JC, Jazayeri A, Khan S, Tehan B, Weir M, Wiggin GR, Marshall FH. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature. 2014 Jul 31;511(7511):557-62. doi: 10.1038/nature13396. Epub 2014 Jul 6. PMID:25042998 doi:http://dx.doi.org/10.1038/nature13396
- ↑ Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013 Feb 14;494(7436):185-94. doi: 10.1038/nature11896. PMID:23407534 doi:http://dx.doi.org/10.1038/nature11896
- ↑ Pin JP, Galvez T, Prezeau L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol Ther. 2003 Jun;98(3):325-54. PMID:12782243
- ↑ Shigemoto R, Nomura S, Ohishi H, Sugihara H, Nakanishi S, Mizuno N. Immunohistochemical localization of a metabotropic glutamate receptor, mGluR5, in the rat brain. Neurosci Lett. 1993 Nov 26;163(1):53-7. PMID:8295733
- ↑ Li G, Jorgensen M, Campbell BM. Metabotropic glutamate receptor 5-negative allosteric modulators for the treatment of psychiatric and neurological disorders (2009-July 2013). Pharm Pat Anal. 2013 Nov;2(6):767-802. doi: 10.4155/ppa.13.58. PMID:24237242 doi:http://dx.doi.org/10.4155/ppa.13.58