Sialyltransferase
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
== Structural highlights == | == Structural highlights == | ||
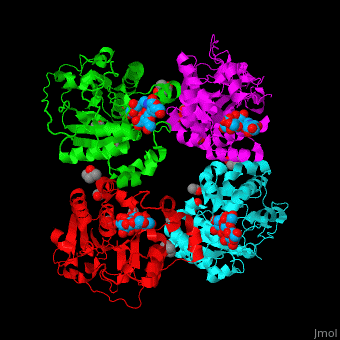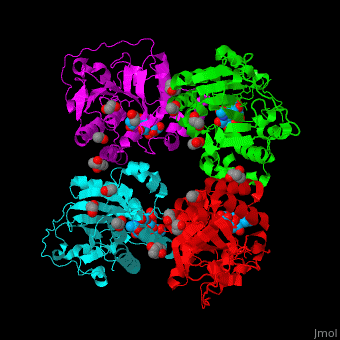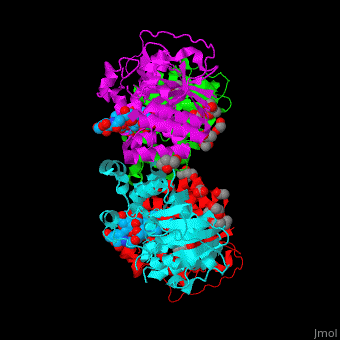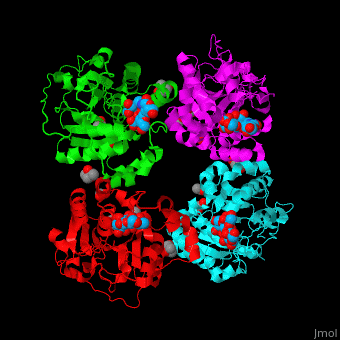
| - | The biological assembly of α-2,3-sialyltransferase is <scene name='74/748880/Cv/2'>homotetramer</scene> ([[2x61]]). The trisaccharide acceptor binds to the open cleft at the C-terminal of SIT<ref>PMID:21832050</ref>. | + | The biological assembly of α-2,3-sialyltransferase is <scene name='74/748880/Cv/2'>homotetramer</scene> ([[2x61]]). The trisaccharide acceptor binds to the open cleft at the C-terminal of SIT<ref>PMID:21832050</ref>. <scene name='74/748880/Cv/3'>Trisaccharide binding site</scene> |
</StructureSection> | </StructureSection> | ||
Revision as of 15:10, 22 December 2016
| |||||||||||
3D structures of sialyltransferase
Updated on 22-December-2016
References
- ↑ Schwartz-Albiez R, Merling A, Martin S, Haas R, Gross HJ. Cell surface sialylation and ecto-sialyltransferase activity of human CD34 progenitors from peripheral blood and bone marrow. Glycoconj J. 2004;21(8-9):451-9. PMID:15750786 doi:http://dx.doi.org/10.1007/s10719-004-5535-5
- ↑ Wang L, Liu Y, Wu L, Sun XL. Sialyltransferase inhibition and recent advances. Biochim Biophys Acta. 2016 Jan;1864(1):143-53. doi: 10.1016/j.bbapap.2015.07.007., Epub 2015 Jul 18. PMID:26192491 doi:http://dx.doi.org/10.1016/j.bbapap.2015.07.007
- ↑ Lee HJ, Lairson LL, Rich JR, Lameignere E, Wakarchuk WW, Withers SG, Strynadka NC. Structural and kinetic analysis of substrate binding to the sialyltransferase CST-II from Campylobacter Jejuni. J Biol Chem. 2011 Aug 8. PMID:21832050 doi:10.1074/jbc.M111.261172