SARS-CoV-2 protein N
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
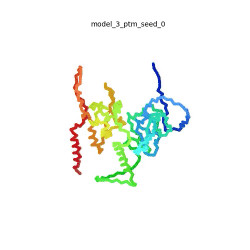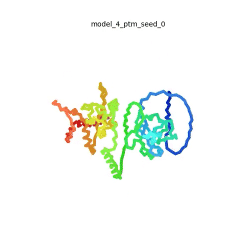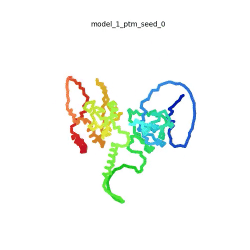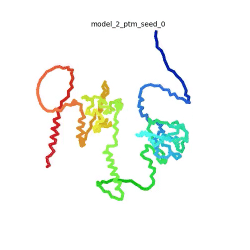
[[Image:SARSCoV2_protein_N_250.gif|250px|left|thumb|'''Morph''' of the top 5 ranked [[AlphaFold]]2 models of '''SARS-CoV-2 Protein N''', ''rainbow'' color coded N-C<ref name="MIT_ColabFold"/>.]]<br />The N-protein consists of 419 amino acids and can be divided into two folded domains and three disordered regions. The three disordered regions dynamically change their conformation and are very flexible (see '''morph''' of the top 5 ranked AlphaFold2 models, to the left, that gives a feel for the flexibility of these domains). This flexibility allows them to rotate and enable the binding to macromolecules like RNA. The two folded domains on the other hand are well structured and have thus been modelled using X-Ray diffraction<ref name="Kang"> PMID: 32363136 </ref> and NMR. <ref name="Savastano">PMID: 32363136 </ref><ref name="Cubuk">PMID:33782395</ref>. | [[Image:SARSCoV2_protein_N_250.gif|250px|left|thumb|'''Morph''' of the top 5 ranked [[AlphaFold]]2 models of '''SARS-CoV-2 Protein N''', ''rainbow'' color coded N-C<ref name="MIT_ColabFold"/>.]]<br />The N-protein consists of 419 amino acids and can be divided into two folded domains and three disordered regions. The three disordered regions dynamically change their conformation and are very flexible (see '''morph''' of the top 5 ranked AlphaFold2 models, to the left, that gives a feel for the flexibility of these domains). This flexibility allows them to rotate and enable the binding to macromolecules like RNA. The two folded domains on the other hand are well structured and have thus been modelled using X-Ray diffraction<ref name="Kang"> PMID: 32363136 </ref> and NMR. <ref name="Savastano">PMID: 32363136 </ref><ref name="Cubuk">PMID:33782395</ref>. | ||
=== N-terminal flexible arm === | === N-terminal flexible arm === | ||
| - | The N-terminal flexible arm is one of the disordered regions, but with parts of transient helicity. Its conformation is significantly affected by the neighbouring folded RNA binding domain (RBD), which reduces the accessible space of the flexible arm and hence supports an expanded configuration of this domain. Interactions with the RBD through fuzzy interactions cause kinetic traps for certain transient configurations. Furthermore, some attractive and repulsive interactions with the RBD are supported by the arginine-rich region (residues 31 - 41) and by residue Phe 17 of the NTD. The arginine-rich motif was found to form a transient alpha helix (H2) | + | The N-terminal flexible arm is one of the disordered regions, but with parts of transient helicity. Its conformation is significantly affected by the neighbouring folded RNA binding domain (RBD), which reduces the accessible space of the flexible arm and hence supports an expanded configuration of this domain. Interactions with the RBD through fuzzy interactions cause kinetic traps for certain transient configurations. Furthermore, some attractive and repulsive interactions with the RBD are supported by the arginine-rich region (residues 31 - 41) and by residue Phe 17 of the NTD. The arginine-rich motif was found to form a transient alpha helix (H2)<ref name="Cubuk"/>. |
=== C-terminal flexible tail: === | === C-terminal flexible tail: === | ||
| - | The disordered C-terminal flexible tail incorporates two more transient helices (H5 and H6). Helix H6 is amphipathic with a hydrophobic face and a positively charged inside. It is probably more highly populated than helix H5. The residues of helix H6 also contribute extensively to an intramolecular interaction with the C-terminal dimerization domain. Thus, helix-formation is in a constant competition with intramolecular interaction with the C-terminal dimerization domain. Experiments revealed “transient but non-negligible interactions [of the CTD] with the dimerization domain” | + | The disordered C-terminal flexible tail incorporates two more transient helices (H5 and H6). Helix H6 is amphipathic with a hydrophobic face and a positively charged inside. It is probably more highly populated than helix H5. The residues of helix H6 also contribute extensively to an intramolecular interaction with the C-terminal dimerization domain. Thus, helix-formation is in a constant competition with intramolecular interaction with the C-terminal dimerization domain. Experiments revealed “transient but non-negligible interactions [of the CTD] with the dimerization domain”<ref name="Cubuk"/>. |
=== Central Linker region (LKR) === | === Central Linker region (LKR) === | ||
| - | The Central Linker region (LKR) is a sequence of polar and charged amino acids within a serine-arginine rich motif and a low number of residues causing steric effects. The resulting electrostatic repulsion of the positively charged residues and the high flexibility due to low steric effects prevents a well structured conformation and causes the disorder. Nevertheless, the region still allows transient structure formation by forming two transient helices: a serine-arginine rich transient helix (H3) and a hydrophobic helix (H4). By measuring the rearrangement time, it has been found that the linker does not interact with the neighbouring folded domains | + | The Central Linker region (LKR) is a sequence of polar and charged amino acids within a serine-arginine rich motif and a low number of residues causing steric effects. The resulting electrostatic repulsion of the positively charged residues and the high flexibility due to low steric effects prevents a well structured conformation and causes the disorder. Nevertheless, the region still allows transient structure formation by forming two transient helices: a serine-arginine rich transient helix (H3) and a hydrophobic helix (H4). By measuring the rearrangement time, it has been found that the linker does not interact with the neighbouring folded domains<ref name="Cubuk"/>. |
| - | The serine-arginine rich motif also provides several putative phosphorylation sites. These sites may regulate protein functions and interactions between membrane proteins and the N-Proteins | + | The serine-arginine rich motif also provides several putative phosphorylation sites. These sites may regulate protein functions and interactions between membrane proteins and the N-Proteins<ref name="Chang"/>. |
| - | The remaining two domains are well organized and represent the majority of the protein. They also contribute the most to the protein’s functions | + | The remaining two domains are well organized and represent the majority of the protein. They also contribute the most to the protein’s functions<ref name="Cubuk"/>. |
=== N-terminal RNA binding domain (RBD): === | === N-terminal RNA binding domain (RBD): === | ||
| - | The β-sheet core has five antiparallel β-strands with a β-hairpin between β2 and β5 and a short helix before β2. The β-hairpin is flexible and may undergo conformational changes during RNA binding. <ref name="Chang"/> Nevertheless, all of the N-protein’s domains and regions are involved in the RNA binding process, which is its main function | + | The β-sheet core has five antiparallel β-strands with a β-hairpin between β2 and β5 and a short helix before β2. The β-hairpin is flexible and may undergo conformational changes during RNA binding. <ref name="Chang"/> Nevertheless, all of the N-protein’s domains and regions are involved in the RNA binding process, which is its main function<ref name="Cubuk"/>. |
=== C-terminal dimerization domain: === | === C-terminal dimerization domain: === | ||
| - | The dimerization domain has more short helices than the RBD. It incorporates eight α helices and only two β-strands which are antiparallel and forming a β-hairpin. The domain is involved in the dimerization (and oligomerization) process, which makes the N-protein functional | + | The dimerization domain has more short helices than the RBD. It incorporates eight α helices and only two β-strands which are antiparallel and forming a β-hairpin. The domain is involved in the dimerization (and oligomerization) process, which makes the N-protein functional<ref name="Chang"/>. |
Revision as of 14:27, 4 February 2022
SARS CoV-2 Protein N
| |||||||||||