SARS-CoV-2 protein NSP11
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
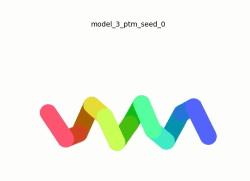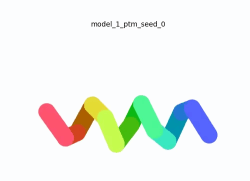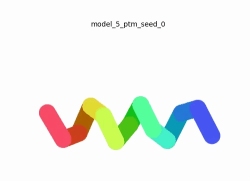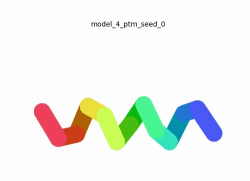
To the right is an '''[[AlphaFold]]2''' 3D model of SARS CoV-2 Protein NSP11 (length = 13 amino acids, i.e. '''SADAQSFLNGFAV''' color coded by the pLDDT scores. It corresponds to the highest ranked model in terms of the pLDDT confidence scores, ''i.e.'', model 5<ref name="MIT_ColabFold">[https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/beta/AlphaFold2_advanced.ipynb MIT ColabFold]</ref>. | To the right is an '''[[AlphaFold]]2''' 3D model of SARS CoV-2 Protein NSP11 (length = 13 amino acids, i.e. '''SADAQSFLNGFAV''' color coded by the pLDDT scores. It corresponds to the highest ranked model in terms of the pLDDT confidence scores, ''i.e.'', model 5<ref name="MIT_ColabFold">[https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/beta/AlphaFold2_advanced.ipynb MIT ColabFold]</ref>. | ||
[[Image:NSP11_AlphaFold2_Movie.gif|250px|left|thumb|'''Morph''' of the top 5 ranked [[AlphaFold]]2 models of '''SARS-CoV-2 Protein NSP11''', ''rainbow'' color coded N-C (blue to red)<ref name="MIT_ColabFold"/>.]] | [[Image:NSP11_AlphaFold2_Movie.gif|250px|left|thumb|'''Morph''' of the top 5 ranked [[AlphaFold]]2 models of '''SARS-CoV-2 Protein NSP11''', ''rainbow'' color coded N-C (blue to red)<ref name="MIT_ColabFold"/>.]] | ||
| + | |||
| + | |||
== Function == | == Function == | ||
'''Non-structural protein 11 (NSP11)''' is the 13 amino acid peptide at the C-term of the large SARS-CoV-2 polyprotein ORF1ab (pp1a). It is the cleavage product of pp1a polyprotein by 3CLpro/Mpro protease at the nsp10/11 junction. Its function and whether it is actually expressed is not known. | '''Non-structural protein 11 (NSP11)''' is the 13 amino acid peptide at the C-term of the large SARS-CoV-2 polyprotein ORF1ab (pp1a). It is the cleavage product of pp1a polyprotein by 3CLpro/Mpro protease at the nsp10/11 junction. Its function and whether it is actually expressed is not known. | ||
Revision as of 15:34, 8 February 2022
| |||||||||||
References
- ↑ 1.0 1.1 MIT ColabFold
- ↑ Gadhave K, Kumar P, Kumar A, Bhardwaj T, Garg N, Giri R. Conformational dynamics of 13 amino acids long NSP11 of SARS-CoV-2 under membrane mimetics and different solvent conditions. Microb Pathog. 2021 Sep;158:105041. doi: 10.1016/j.micpath.2021.105041. Epub 2021, Jun 10. PMID:34119626 doi:http://dx.doi.org/10.1016/j.micpath.2021.105041