User:Eduardo Soares/Sandbox 1
From Proteopedia
(Difference between revisions)
m |
|||
| Line 1: | Line 1: | ||
=VCA0042/plzD complexed with c-di-GMP= | =VCA0042/plzD complexed with c-di-GMP= | ||
==Introduction== | ==Introduction== | ||
| - | The PDB code 2RDE represents a PilZ protein complexed with cyclic diguanylate monophosphate (c-di-GMP) there is a molecular interaction involved in the regulation of bacterial biofilm formation and motility. The complex is formed by the binding of the small signaling molecule c-di-GMP to a protein domain known as PilZ. This interaction plays a crucial role in bacterial physiology and is implicated in various cellular processes. | + | The PDB code 2RDE represents a PilZ protein complexed with cyclic diguanylate monophosphate (c-di-GMP) in ''Vibrio cholera'' there is a molecular interaction involved in the regulation of bacterial biofilm formation and motility. The complex is formed by the binding of the small signaling molecule c-di-GMP to a protein domain known as PilZ. This interaction plays a crucial role in bacterial physiology and is implicated in various cellular processes. |
<StructureSection load='2rde' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='2rde' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| Line 19: | Line 19: | ||
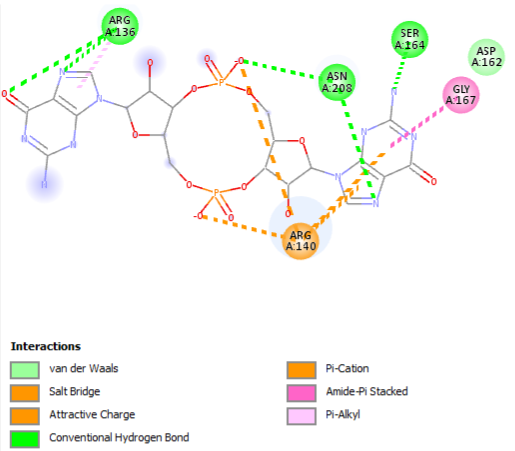
The binding of c-di-GMP to the PilZ domain is specific and occurs through conserved amino acid residues within the domain. The affinity and specificity of the interaction can vary among different PilZ-containing proteins, allowing for fine-tuning of c-di-GMP signaling in different bacterial species. | The binding of c-di-GMP to the PilZ domain is specific and occurs through conserved amino acid residues within the domain. The affinity and specificity of the interaction can vary among different PilZ-containing proteins, allowing for fine-tuning of c-di-GMP signaling in different bacterial species. | ||
| - | == Function == | + | [[Image:Diagrama 2D.png]] |
| + | |||
| + | == Function in Virio cholera == | ||
| + | |||
| + | Cyclic diguanylate monophosphate (c-di-GMP) signaling plays a crucial role in the life cycle of Vibrio cholerae, a bacterium responsible for causing the human disease cholera (Islam et al., 1993). Cholera is characterized by potentially life-threatening diarrhea resulting from the secretion of cholera toxin. Studies have revealed that c-di-GMP regulates the expression of virulence genes in V. cholerae (Tischler and Camilli, 2005). Among the proteins involved in c-di-GMP signaling is VieA, a c-di-GMP phosphodiesterase that modulates cholera toxin production by influencing the cytosolic levels of c-di-GMP. Additionally, the genome of V. cholerae contains five PilZ domain-containing proteins, one of which is VCA0042. These proteins diverged in sequence before the divergence of the Vibrio lineage (Figure 1A) and are believed to function as regulators of various cellular processes controlled by c-di-GMP. VCA0042 has recently been named PlzD and has been shown to bind c-di-GMP when immobilized on nitrocellulose in its native conformation (Pratt et al., 2007). Furthermore, a study has demonstrated that either VCA0042/PlzD or VC2344/PlzC, another PilZ domain-containing protein in V. cholerae, is required for efficient colonization of the intestines of mice by the El Tor biotype of V. cholerae in a virulence assay. | ||
== Relevance == | == Relevance == | ||
Revision as of 12:34, 23 June 2023
VCA0042/plzD complexed with c-di-GMP
Introduction
The PDB code 2RDE represents a PilZ protein complexed with cyclic diguanylate monophosphate (c-di-GMP) in Vibrio cholera there is a molecular interaction involved in the regulation of bacterial biofilm formation and motility. The complex is formed by the binding of the small signaling molecule c-di-GMP to a protein domain known as PilZ. This interaction plays a crucial role in bacterial physiology and is implicated in various cellular processes.
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644