User:Eduardo Soares/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
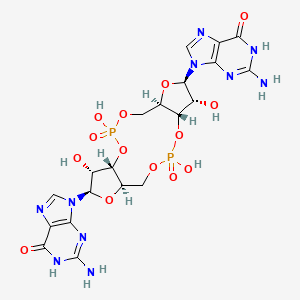
The synthesis of c-di-GMP involves the conversion of two guanosine triphosphate (GTP) molecules by diguanylate cyclases (DGCs), while its breakdown yields 5′-phosphoguanylyl-(3′-5′)-guanosine (pGpG) through the action of specific phosphodiesterases (PDEs). pGpG is subsequently cleaved into two guanosine monophosphate (GMP) molecules (Figure 1). DGC activity is associated with the GGDEF domain, named after the crucial amino acid sequence motif found in the enzyme's active site. On the other hand, c-di-GMP-specific PDE activity is linked to the EAL or HD-GYP domains, with their respective amino acid motifs playing essential roles in their enzymatic functions. | The synthesis of c-di-GMP involves the conversion of two guanosine triphosphate (GTP) molecules by diguanylate cyclases (DGCs), while its breakdown yields 5′-phosphoguanylyl-(3′-5′)-guanosine (pGpG) through the action of specific phosphodiesterases (PDEs). pGpG is subsequently cleaved into two guanosine monophosphate (GMP) molecules (Figure 1). DGC activity is associated with the GGDEF domain, named after the crucial amino acid sequence motif found in the enzyme's active site. On the other hand, c-di-GMP-specific PDE activity is linked to the EAL or HD-GYP domains, with their respective amino acid motifs playing essential roles in their enzymatic functions. | ||
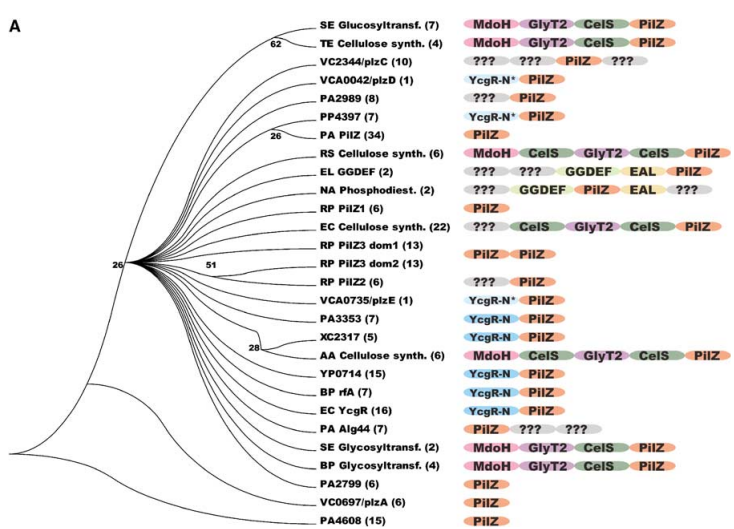
The significance of c-di-GMP regulatory systems gained considerable attention following the advent of whole genome sequencing, revealing the widespread presence of GGDEF and EAL domains in bacterial species, often with a large number of encoded proteins. Artificial modulation of cellular c-di-GMP levels through overproduction of GGDEF domain proteins led to enhanced adhesin and biofilm matrix component synthesis, while impairing motility and acute virulence functions. Conversely, overproduction of EAL domain proteins produced opposite phenotypes (Figure 1). Current research aims to assign specific molecular functions to GGDEF, EAL, and HD-GYP domain proteins and unravel their integration into complex regulatory networks. Notably, recent findings have identified biologically active GGDEF and EAL domain proteins lacking DGC or PDE activity, exemplifying their involvement in transcriptional regulation and RNA degradation. | The significance of c-di-GMP regulatory systems gained considerable attention following the advent of whole genome sequencing, revealing the widespread presence of GGDEF and EAL domains in bacterial species, often with a large number of encoded proteins. Artificial modulation of cellular c-di-GMP levels through overproduction of GGDEF domain proteins led to enhanced adhesin and biofilm matrix component synthesis, while impairing motility and acute virulence functions. Conversely, overproduction of EAL domain proteins produced opposite phenotypes (Figure 1). Current research aims to assign specific molecular functions to GGDEF, EAL, and HD-GYP domain proteins and unravel their integration into complex regulatory networks. Notably, recent findings have identified biologically active GGDEF and EAL domain proteins lacking DGC or PDE activity, exemplifying their involvement in transcriptional regulation and RNA degradation. | ||
| - | |||
| - | [[Image:Figura2.jpg]] <ref>Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7, 263–273 (2009). https://doi.org/10.1038/nrmicro2109<\ref> | ||
| - | |||
==Molecular Interaction== | ==Molecular Interaction== | ||
Revision as of 09:17, 26 June 2023
VCA0042/plzD complexed with c-di-GMP
Introduction
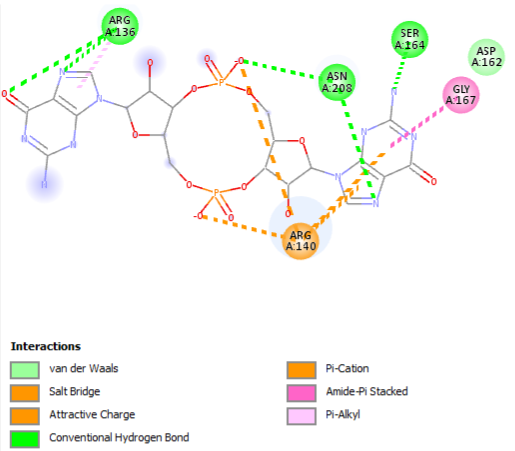
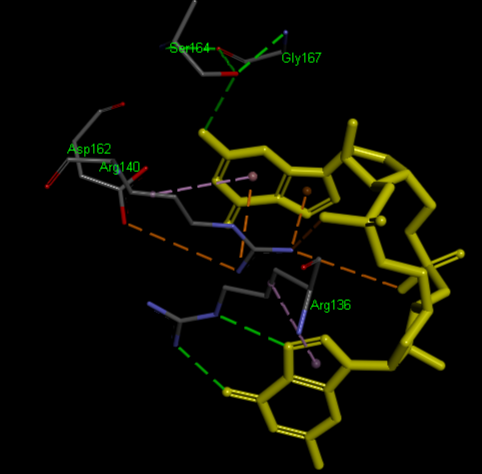
The PDB code 2RDE represents a PilZ protein complexed with cyclic diguanylate monophosphate (c-di-GMP) in Vibrio cholera there is a molecular interaction involved in the regulation of bacterial biofilm formation and motility. The complex is formed by the binding of the small signaling molecule c-di-GMP to a protein domain known as PilZ. This interaction plays a crucial role in bacterial physiology and is implicated in various cellular processes. [1]
| |||||||||||
References
- ↑ Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153-66. doi: 10.1038/sj.emboj.7601918. Epub 2007 Nov 22. PMID: 18034161; PMCID: PMC2140105.
- ↑ Pecina, Anna, et al. The Stand-Alone PilZ-Domain Protein MotL Specifically Regulates the Activity of the Secondary Lateral Flagellar System in Shewanella putrefaciens. Frontiers in Microbiology, vol. 12, 2021. Frontiers, https://www.frontiersin.org/articles/10.3389/fmicb.2021.668892.
- ↑ Bena
 ch J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153-66. doi: 10.1038/sj.emboj.7601918. Epub 2007 Nov 22. PMID: 18034161; PMCID: PMC2140105.
ch J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153-66. doi: 10.1038/sj.emboj.7601918. Epub 2007 Nov 22. PMID: 18034161; PMCID: PMC2140105. - ↑ Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153-66. doi: 10.1038/sj.emboj.7601918. Epub 2007 Nov 22. PMID: 18034161; PMCID: PMC2140105.
- ↑ Purificação, Aline Dias da, et al. The World of Cyclic Dinucleotides in Bacterial Behavior. Molecules, vol. 25, n. 10, Janeiro de 2020, p. 2462. www.mdpi.com, https://doi.org/10.3390/molecules25102462.
- ↑ Purificação, Aline Dias da, et al. The World of Cyclic Dinucleotides in Bacterial Behavior. Molecules, vol. 25, n. 10, Janeiro de 2020, p. 2462. www.mdpi.com, https://doi.org/10.3390/molecules25102462.
- ↑ Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153-66. doi: 10.1038/sj.emboj.7601918. Epub 2007 Nov 22. PMID: 18034161; PMCID: PMC2140105.