MOLECULAR MODEL:
The initial view here is a ball-and-stick representation of the molecular structure of myoglobin.
SECONDARY STRUCTURE:
This next view simplifies things, and just shows a of the secondary structure of the protein.
You see how the that maintain the main secondary structure of the protein are arranged in this next view.
THE GLOBIN FOLD:
In this next view, the eight are each coloured differently. This gives you an impression of the classic globion fold. The α-helices pack together tightly, and there is very little space in the centre of the protein.
HYDROPHOBICITY:
Globular folds like this are characterised by a polar, , which interacts with the aqueous solvent, and a hydrophobic core.
Hydrophobic, Polar
The next view shows a section through the protein that highlights the better.
This view has been produced in the software by a process known as 'slabbing'. You can still rotate the molecule around - whatever view you see will the the front part of the view of the protein cut off.
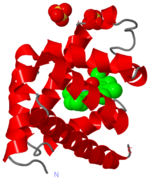
THE HEME GROUP:
Now let's turn our attention to the main function of myoglobin - oxygen binding.
Oxygen is bound by a , which sits in a hydrophobic pocket in the myoglobin protein.
Central to the heme group is an .
PROXIMAL AND DISTAL HISTIDINES: The iron atom sits either side of the side chains of two .
One of these (coloured cyan) is attached to the iron atom, and is known as the proximal histidine. The other (green) is called the distal histidine.
OXYGEN:
The space between the iron and the distal histidine is where the (pink) binds.