This is a default text for your page Scott H. Vanson/Sandbox 1. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
General Description
Human polymerase θ (pol θ) is large, 290kD enzyme consisting of three distinct domains [3][4]. An N-terminal helicase-like domain, whose exact cellular functions are a topic of on-going debate and research[5][6], is linked to a C-terminal, family A DNA polymerase domain by a large and disordered central region[4]. Notably, pol θ is the only known human polymerase to contain a polymerase and helicase domain in one molecule[7]. Crystal structures have been solved for the apo form of the helicase-like domain and the ternary complex of the polymerase domain. The focus of this wiki is the polymerase domain.
Pol θ is thought to promote overall genomic stability by performing several distinct cellular functions. The primary role of the enzyme is to repair of double-stranded DNA breaks as the key enzyme in an error-prone non-homologous end-joining pathway called alternative end-joining[8] or theta-mediated end-joining. Other functions include translesion synthesis, the ability of the polymerase to bypass and extend past a site of oxidative DNA damage[9], base excision repair [10], and possibly DNA replication timing [11]. Pol θ has the specialized ability to extend DNA from minimally-paired primers (termed microhomologous)Template:Cn. Repair by this enzyme is considered to error-prone due to its tendency to add or delete short indels [8].
Several types of cancer, such as breast, ovarian, and oral carcinomas, have shown significantly higher expression levels of pol θ and correlate to poorer patient outcomes[12][13][14]. Genomic studies have shown that more than half of epithelial ovarian cancers have defects in the error-free repair pathway of homologous recombination[15] and, as a result, have an increased dependence on theta-mediated end-joining [12]. Double-stranded break repair by pol θ may be thought of as a "backup" pathway which cells depend on more when the machinery involved in homologous recombination is compromised or otherwise unavailable. This enzyme has been identified as a potential therapeutic target due to overexpression in cancers in combination with studies that have shown inhibition of pol θ to sensitize human and mouse cells to radiation and chemical agents which induce double-stranded breaks[12][16][17].
Structural Highlights
Two crystal structures of the polymerase domain have been solved bound, inserting ddATP opposite tetrahydrofuran (THF, representing an abasic site) and inserting ddGTP opposite dCMP[7]. An overall assessment of the structures display the canonical . The DNA is thought to "sit" on the palm and is enclosed by the thumb and fingers which contact the minor groove. The closed conformation of the polymerase domain becomes obvious when compared to the , such as Thermus aquaticus DNA polymerase I.
ddATP Opposite THF
An inspection of the active site reveals the , in this instance calcium. This structure required the use of calcium, a known inhibitor of polymerase activity, as the primer strand retains the 3' hydroxyl which would otherwise be subjected to nucleophilic attack. The similarly conserved .
θθ
dCMP Opposite ddGTP
from the same study was solved with magnesium as the coordinated metal, a 27 amino acid N-terminal truncation, and a blunted DNA oligomer to remove the 3' template overhang. The overall structure is virtually identical to the aforementioned structure complexed with calcium, with the exception of a slight shift in position of the O-helix to be closer to the cognate C:G basepair.
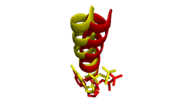
Alignment of O-helices and incoming nucleotides. Mg complex (yellow) shifts slightly closer to incoming ddGTP than Ca complex (red) relative to its incoming ddATP.
Structural Insights into Function
Proofreading Activity
Family A DNA polymerases harbor an N-terminal exonuclease domain and generally conserve the overall fold. Some members of this family, e.g. E. coli pol I, display active 3'-5' proofreading activity via . Other members, such as pol θ, do not harbor proofreading activity[7] and at equivalent positions differ in identity and arrangement.
Translesion Synthesis
An R2254V variant was made to investigate the importance of this residue in pol θ's ability to extend single-stranded DNA and bypass abasic sites and bulky thymine glycol lesions.
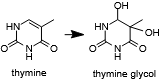
Thymine may oxidized to form a bulky lesion that must be repaired or bypassed.
This mutant retained its ability to extend double-stranded but not single-stranded DNA and also was not able to bypass abasic sites or thymine glycol. These findings indicated to the authors that the salt bridge between R2254 and the 3'-terminal phosphate of the primer is required to compensate for the missing contacts to the template strand due to a lesion.
Related Proteins


