Hormone-sensitive lipase
Introduction
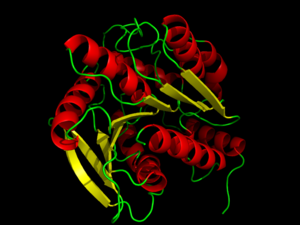
Hormone-Sensitive Lipase from
3dnm. Alpha helices and beta sheets are shown in red and yellow, respectively.
Hormone-sensitive lipases (HSL) represent a class of esterases within the α/β hydrolase family. HSL catalyzes the cleavage of ester bonds in fatty acid molecules when stimulated by a hormone. [1] The activation and mobilization of hormone-sensitive lipase can be triggered by various catecholamines and inhibited by insulin. [2] Catecholamines, such as epinephrine, are rapidly spread throughout the body during times of energy mobilization, like in the fight of flight response. Conversely, insulin triggers glucose uptake, requiring the storage of energy, opposing HSL's function. HSL is clinically relevant, because the mobilization of or inability to mobilize fats in cells is directly related to fat accumulation seen in artherosclerosisand obesity.[3] Such diseases are characterized by an accumulation of fats and researchers are investigating whether HSL's activity plays a role or not. [2] Investigation of HSL's structure and function could provide a better clinical understanding of these diseases. [3]
Activation of HSL
Briefly, HSL is stimulated by the binding of catecholamines to β-adrenergic receptors. β-adrenergic receptors coupled with adenylate cyclase (AC) then stimulate G-proteins to increase the levels of cystolic cAMP. Elevated levels of cAMP leads to an activation protein kinase A (PKA) leading to phosphorylation of serine residues on HSL activating and translocating HSL to lipid droplets for lipolysis. Conversely, insulin signaling decreases cystolic cAMP levels, resulting in a decreased HSL mobilization. [1]
Structure of hormone-sensitive lipase
are generally well-conserved across domains, including prokaryotes, showing 29, 26, and 22% residue overlap in Alicyclobacillus acidocaldarius, Archaeoglobus fulgidus, and Bacillus subtilis, respectively. [4] HSL is composed of two main structural domains, consisting of a slightly variable N-terminus (shown in blue in the ) that is thought to contribute to numerous factors including activity, specificity, regioselectivity, thermophilicity, and thermostability. [4] Research speculates that the N-terminal domain, consisting of about 300 residues, mediates protein-protein interactions, and possibly subsequent lipid binding. [3] The second domain of HSL is the C-terminal catalytic domain (colors other than blue), which contains serine residue phosphorylation sites as well as the catalytic triad, viewed , a charge relay network that is characteristic of many hydrolases, such as chymotrypsin. [3] With respect to sequence conservation across species, it has been shown that the catalytic domain, including the triad, is conserved across domains, but the domain containing the N-terminus shows little conservation. [4] Size-exclusion chromatography studies have shown that HSL has a that is approximately 16Å deep, suggesting that HSL primarily hydrolyzes shorter chained molecules. [4]
Catalytic triad
The catalytic triad toward the middle of HSL. The catalytic triad is composed of residues . The Ser157 residue sits at a site deemed the "nucleophilic elbow," that models an approximate torsion of Φ = 60° and Ψ =-120°. This nucleophilic elbow is stabilized by a hydrogen bond between the proximal nitrogen and oxygen atoms of His281 and Glu251, respectively. This model also shows the strong nucleophilic character of Ser157, portraying the covalent binding to . Return to default view, .
Inhibition of hormone-sensitive lipase
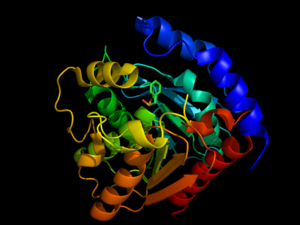
Hormone-Sensitive Lipase Complex with PMSF from
3h17Hormone-sensitive lipase can be inhibited by phenylmethylsufonyl flouride (PMSF) entering into the . The experiments performed to test this inhibition used different strains of bacteria, which had a different order of residues but the same catalytic effect. This experiment tested the Ser144 residue rather than the Ser157 residue seen in other HSL proteins. PMSF inhibits enzymes by binding to the Ser144 residue of the serine protease active sites so that the normal catalytic activity cannot be carried out. This inhibitor will only bind to the active site Ser144 because of its participation in the charge relay of the . This hyper activity allows the sulfonyl group of PMSF to to the Ser144 residue to disrupt its activity. Because of this Ser144 residue specificity, PMSF does not inhibit all kinds of lipases, just those dependent on Ser144 residues in the active site. PMSF is highly degradable in aqueous solutions so it does not inhibit for very long periods of time in its natural environment. PMSF binding induces only a minor conformational change from the native protein.[5]
Inhibition or decreased activity of hormone-sensitive lipases can possibly lead to disorders such as atherosclerosis, obesity, and type 2 diabetes. Inability to break down fatty acid molecules due to inactive hormone-sensitive lipases has been reported in many cases of obesity and type 2 diabetes. Increased inhibition of HSL by PMSF could, in theory, also be a possible cause of these diseases.[6]


