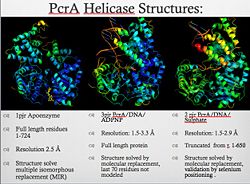
PcrA helicase
From Proteopedia
Contents |
What is a Helicase?
| |||||||||
| 1pjr, resolution 2.50Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene: | PCRA (Geobacillus stearothermophilus) | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Helicases are nucleic acid–dependent ATP-ases that are capable of unwinding DNA [1] or RNA [2] duplex substrates. As a consequence, they play roles in almost every process in cells that involves nucleic acids, including DNA replication and repair, transcription, translation, ribosome synthesis (1).
PcrA a Simple Model for Helicases
PcrA is part of the replication machinery of the Geobacillus stearothermophilusa gram (+) bacteria, This helicase is part of the superfamily I of Helicases. monomeric protein that is mainly . This helicase was reported as a mutation in the gen PcrA from "Stapphylococcus aerous", this mutation was related to a deficiency in the replication of a reporter plasmid.[3]
PcrA Biochemistry
PcrA is has an ATPas activityt which directionality is from 3' to 5' helicase strand separation reaction. The enzyme shows a specificity for the DNA substrate in gel mobility assays with the preferred substrate being one containing both single and double stranded regions of DNA. In contrast to Rep and UvrD from E. coli, there is not evidence for dimerisation of the enzyme using gel filtration, or by crosslinking in the presence of combinations of Mg2+, nucleotides and DNA. Moreover, kcat for ATP hydrolysis is constant over a large range of protein concentrations. Therefore, the protein appears to be monomeric under all conditions tested, including in the structure of two crystal forms of PcrA.[4]
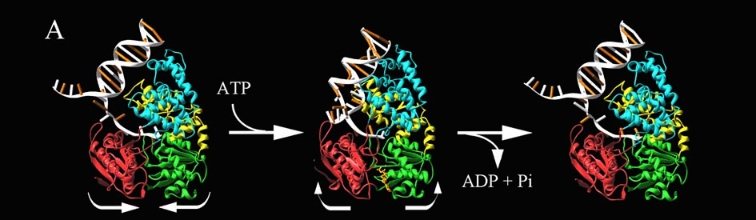
PcrA Helicase Mechanism : The Mexican Wave
Professor Dale B. Wigley' group in 1996-1999 was able to crystalize the intermediate states from PcrA, giving solution to the controversy of what kind of mechanism this helicase has. [5] Two crystal form of the enzyma, one couple with a 10 mer DNA and a non hydrolizable form of ATP (ATPnP) (pdb id: 3pjr 3pjr, and another a truncated form embebed in sulfate (pdb id: 2pjr 2pjr), give a light in a model for how ATP hydrolysis results in motor movement along ssDNA. In the figure below step 1 (top) is the ATP free (product) ssDNA conformation. The DNA bases are labelled arbitrarily. On binding ATP, F626 creates a new binding pocket for base 6. Likewise, F64 destroys an acceptor pocket for base 2, forcing it to move to the position occupied by base 1. After ATP hydrolysis, the grip on base 6 is released. When the Y257 pocket is re-opened due to movement of F64, bases 3-6 can now flip through the acceptor pockets to their new positions. This model predicts that each ATP hydrolysis event will advance PcrA one base along ssDNA.[6]
|
The link below show a movie with the principal characteristics of this protain as long with the inchworm mode. Pcr4 Helicase and Mexican Wave
The Superfamily 1 (SF1)
PcrA share structural domains with the Rec helicases, like UvrD and RepD from E. coli
| |||||||||
| 2is1, resolution 2.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | uvrD (Escherichia coli) | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
| |||||||||
| 1uaa, resolution 3.00Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Structure of the lac repressor
|
The lac repressor protein ( showing chain A in 1lbg, resolution 4.8 Å), starting at the N-terminus, begins with a DNA-binding "headpiece", followed by a hinge region, then an N-terminal ligand-binding subdomain and a C-terminal ligand binding subdomain, a linker, and a C-terminal tetramerization helix[1]. (.) In the absence of DNA, the hinge region does not form the alpha helix shown here.
As can be seen when the chain is
| N | C |
each of the ligand-binding subdomains is made up of two discontinuous segments.
The lac repressor forms . Dimerization buries 2,200 Å2 of surface, including a ,
forming a hydrophobic core (shown with 1lbi, resolution 2.7 Å, lacking the DNA-binding domain due to disorder).
 |
The most highly is the surface that contacts DNA[2]. (Only alpha carbon atoms are shown here, without sidechains, because sidechains were not resolved in the 4.8 Å 1lbg model.) The dimerization surfaces are the of the ligand-binding domains[3]. (This scene shows sidechains, using the 2.7 Å model in 1lbi, which lacks the DNA-binding domain due to disorder.)
<--! scene with translucent :a is #11, but I didn't like it. --> The C-terminal tetramerization helices tether two dimers, and thus the functional form of with two DNA-binding sites.
See Also
- Category: Lac repressor and Category: Lac Repressor, automatically-generated pages that list PDB codes for lac repressor models.
- Morphs where the morph of the lac repressor is used as an example.
References & Notes
- ↑ This domain coloring scheme is adapted from Fig. 6 in the review by Lewis (C. R. Biol. 328:521, 2005). Domains are 1-45, 46-62, (63-162,291-320), (163-290,321-332), 330-339, and 340-357.
- ↑ Conservation results for 1lbg are from the precalculated ConSurf Database, using 103 sequences from Swiss-Prot with an average pairwise distance of 2.4.
- ↑ Conservation results for 1lbi are from the ConSurf Server, using 100 sequences from Uniprot with an average pairwise distance of 1.3.
History of DNA Helicase Discovery
About this Structure
1PJR is a Single protein structure of sequence from Geobacillus stearothermophilus. Full crystallographic information is available from OCA.
Reference
Crystal structure of a DExx box DNA helicase., Subramanya HS, Bird LE, Brannigan JA, Wigley DB, Nature. 1996 Nov 28;384(6607):379-83. PMID:8934527
Page seeded by OCA on Mon Jul 28 01:38:49 2008
Proteopedia Page Contributors and Editors (what is this?)
Luis E Ramirez-Tapia, Michal Harel, Eran Hodis, David Canner