Sandbox Reserved 649
From Proteopedia
| This Sandbox is Reserved from 30/08/2012, through 01/02/2013 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 636 through Sandbox Reserved 685. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://proteopedia.org/w/Help:Getting_Started_in_Proteopedia
Argininosuccinate Lyase
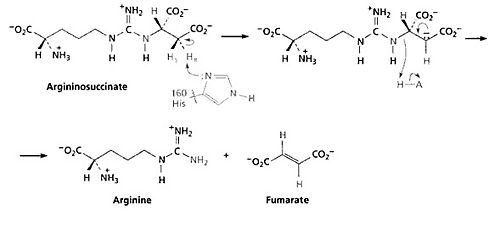
Argininosuccinate Lyase(ASL), also known as Argininosuccinase, is an enzyme that catalyzes the reversible reaction of argininosuccinate to arginine and fumerate. ASL catalyzes the fourth step in the urea cycle taking place in liver cytosol after argininosuccinate synthetase. ASL is part of a superfamily of homotetrameric enzymes that includes δ-crystallin, class II fumarase and aspartase in which a Cα–N or Cα–O bond is cleaved. Among the superfamily, Human ASL is about 64-72% identical to δ-crystallin an avian eye lens protein. The structure of ASL is composed of mostly alpha helices. ASL has been found in E. coli, yeast, rats, humans, and commonly found in birds and reptiles. ASL deficiency is the second most common urea cycle disorder with an occurence of 1 in every 70,000 live births. [1]
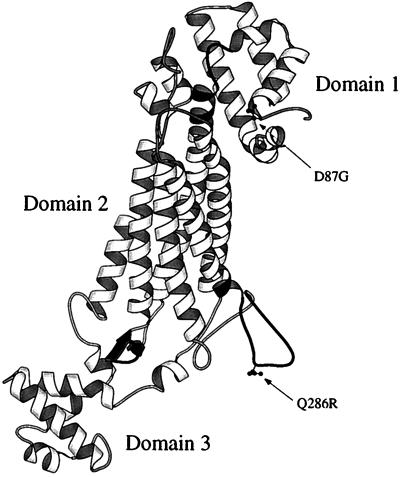
StructureThe structure of human Argininosuccinate lyase was solved in 1997 through x-ray diffraction with a resolution of 4.2 Å. ASL is composed of four single polypeptide monomers each with a molecular weight of 49 to 52 kDa.[2] Each monomer contains three predominantly alpha helical domains in which the first and third have a similar topography of two perpendicular helix-turn-helix motifs. The second domain is characterized by a helical bundle of which three from each monomer interacts to form a dimer. Altogether, ASL is a homotetramer approximately 64% helical structures and 4% beta sheets. Structural studies indicate the is composed of an acid catalyst Ser283 and a catalyic base His162.[3] The residue Lys289 is thought to help stabilize the carbanion intermediate. Studies indicate that a hydroxy group of either threonine or serine should be present at residue 161 to help position Lys289. A is seen in the open conformation with residues 270 to 290 and is used to sequester the substrate argininosuccinate.
MechanismThe fourth step of the urea cycle is catalyzed by argininosuccinate lyase. The products of this reaction are arginine and fumarate. The fumarate product is an important link between the urea cycle and the citric acid cycle. The base initiates the reaction by deprotonating the carbon adjacent to the arginine, or leaving group. The process occurs by an E1cB mechanism with loss of the pro-R hydrogen (the hydrogen that will give the R configuration of a molecule after it is abstracted) and with anti-stereochemistry. The E1cB elimination reaction is a special type of elimination reaction in organic chemistry. This reaction mechanism explains the formation of alkenes from mostly alky halides through a carbanion intermediate given specified reaction condition and specified substrates. There are some evidences have shown that Histidine 162 or Threonine 161 of ASL is responsible for the proton abstraction of the carbon, either directly or indirectly.[5] ImplicationsArgininosuccinate lyase deficiencyArgininosuccinate lyase deficiency is a urea cycle disorder which can develop in the neonatal period or later on in childhood; this is a rare, autosomal recessive disorder. Argininosuccinate lyase is an intermediate enzyme in the urea synthesis pathway and its function is imperative to the continuation of the cycle. As result of this enzyme defect, patients have increased ammonia, argininosuccinate, and citrulline accumulation in the blood.[6] Ammonia builds to toxic levels, resulting in hyperammonnemia and may affect the nervous system and eventually damage the liver. There is biochemical evidence that shows rises in ammonia can inhibit glutaminase and therefore limit the rate of synthesis of neurotransmitters such as glutamate, which could lead to the developmental delay in argininosuccinic aciduria patients.[7] Genetic Factors ASL deficiency is inherited in an autosomal recessive manner. At conception, each child of an affected individual has a 25% chance of being affected, a 50% chance of being an asymptomatic carrier, and a 25% chance of being unaffected and not a carrier. SymptomsNeonatal onset presents in the first 2-3 days of life -vomiting -lethargy -respiratory alkalosis -hypothermia -seizure -coma Late onset -developmental delay -intellectual disability -seizures -skin and hair abnormalities Treatments1)Treatment consists of a low protein diet, arginine supplementation to help complete the urea cycle, ammonia scavenging drugs in some cases and supplemental carnitine if the patients have a secondary deficiency.[8] 2)Liver transplant offers a partial correction of the enzyme deficiency and improved metabolic status. References
|