Peroxisome Proliferator-Activated Receptors
From Proteopedia
| Line 17: | Line 17: | ||
==Natural Ligands== | ==Natural Ligands== | ||
[[Image: Linoleic_Acid.png|400px|left|thumb| PPARγ Ligand, Linoleic Acid]] | [[Image: Linoleic_Acid.png|400px|left|thumb| PPARγ Ligand, Linoleic Acid]] | ||
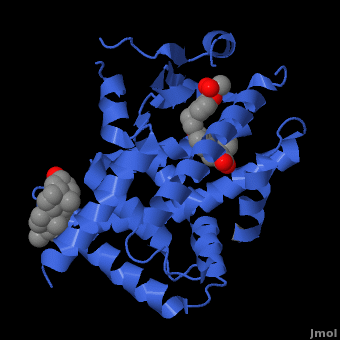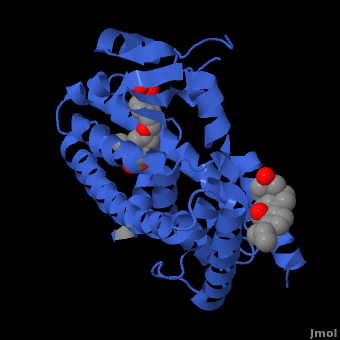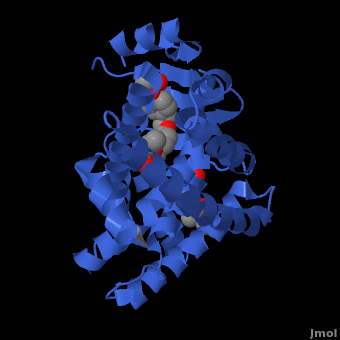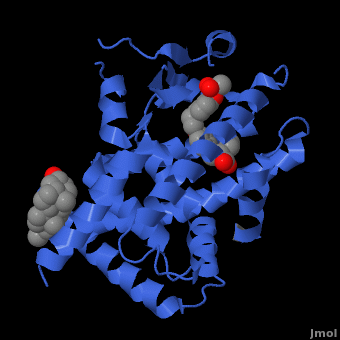
| - | <applet load=" 1i7g2.pdb" size="400" color="white" frame="true" spin="on" Scene ="Peroxisome_Proliferator-Activated_Receptors/Ppar_opening_2/ | + | <applet load=" 1i7g2.pdb" size="400" color="white" frame="true" spin="on" Scene ="Peroxisome_Proliferator-Activated_Receptors/Ppar_opening_2/2" caption="Crystal Structure of Human <scene name='Peroxisome_Proliferator-Activated_Receptors/Ppar_opening_2/2'>PPAR</scene>" align="right"/> |
PPARγ binds polyunsaturated fatty acids like linoleic acid, linolenic acid, and [http://en.wikipedia.org/wiki/Eicosapentaenoic_acid eicosapentaenoic acid] at affinities that are in line with serum levels. PPARα binds a variety of saturated and unsaturated fatty acids including [http://en.wikipedia.org/wiki/Palmitic_acid palmitic acid], [http://en.wikipedia.org/wiki/Oleic_acid oleic acid], [http://en.wikipedia.org/wiki/Linoleic_acid linoleic acid], and arachidonic acid.<ref>PMID:1316614</ref> PPARδs ligand selectivity is intermediate between that of the other isotypes and is activated by palmitic acid and a number of eicosanoids.<ref>PMID:7836471</ref> | PPARγ binds polyunsaturated fatty acids like linoleic acid, linolenic acid, and [http://en.wikipedia.org/wiki/Eicosapentaenoic_acid eicosapentaenoic acid] at affinities that are in line with serum levels. PPARα binds a variety of saturated and unsaturated fatty acids including [http://en.wikipedia.org/wiki/Palmitic_acid palmitic acid], [http://en.wikipedia.org/wiki/Oleic_acid oleic acid], [http://en.wikipedia.org/wiki/Linoleic_acid linoleic acid], and arachidonic acid.<ref>PMID:1316614</ref> PPARδs ligand selectivity is intermediate between that of the other isotypes and is activated by palmitic acid and a number of eicosanoids.<ref>PMID:7836471</ref> | ||
<br /> | <br /> | ||
| Line 25: | Line 25: | ||
==PPAR Structure== | ==PPAR Structure== | ||
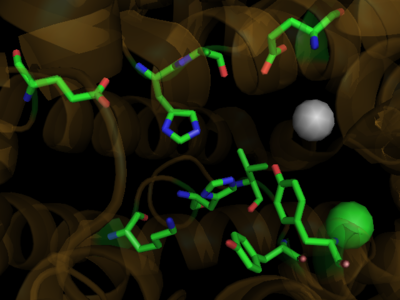
===Ligand Binding Domain=== | ===Ligand Binding Domain=== | ||
| - | The structures of the PPARs are very similar over each isotype. All PPAR isotypes have a ligand binding domain (LBD). The LBD, which is located in the C-terminal half of the receptor, is composed of 13 α-helices and a four-stranded δ-sheet. <scene name='Peroxisome_Proliferator-Activated_Receptors/Ligand_binding_pocket/ | + | The structures of the PPARs are very similar over each isotype. All PPAR isotypes have a ligand binding domain (LBD). The LBD, which is located in the C-terminal half of the receptor, is composed of 13 α-helices and a four-stranded δ-sheet. <scene name='Peroxisome_Proliferator-Activated_Receptors/Ligand_binding_pocket/2'>The ligand binding pocket</scene> is Y-shaped and consists of an <scene name='Peroxisome_Proliferator-Activated_Receptors/Y_shaped/4'>entrance and two pockets, Arm I and Arm II, along with a "charge-clamp"</scene>. The ligand binding pocket of PPARs is quite large in comparison to that of other nuclear receptors (about 1400 cubic angstroms) which allows the PPARs to interact with a broad range of structurally distinct ligands.<ref>PMID:9744270</ref>. Within Arm I, four polar resides are conserved over all PPAR isotypes, <scene name='Peroxisome_Proliferator-Activated_Receptors/4_conserved_residues/1'>namely Ser280, Tyr314, His440, and Tyr464</scene> in the case of PPARα. These residues are part of a hydrogen bonding network that is formed with the carboxylate group of fatty acids and other ligands upon binding.<ref>PMID:16405912</ref> The <scene name='Peroxisome_Proliferator-Activated_Receptors/Helix_h12/4'>ligand-dependent activation domain (AF-2) helix H12</scene>, whose function is to generate the receptors’ co-activator binding pocket is located at the C-terminal end of the LBD.<ref>PMID:11027271</ref> The conserved hydrogen bonding network in <scene name='Peroxisome_Proliferator-Activated_Receptors/Helix_h12_in_place/1'>Arm I also helps hold the AF2-helix in the active conformation</scene>, promoting co-activator binding.<ref>PMID:17317294</ref> <scene name='Peroxisome_Proliferator-Activated_Receptors/Arm_ii_hydrophobic/3'>Arm II is highly hydrophobic </scene>and is thus ideal for binding the hydrophobic tail of fatty acids via Van der Waals interactions. |
Despite over 80% of the ligand binding cavity residues being conserved over all PPAR isotypes, it is the remaining 20% that creates the ligand specificity seen between isotypes. A few examples illustrate this point. In PPARδ, the cavity is significantly narrower adjacent to the AF-2 helix and Arm I. This prevents PPARδ from accommodating large headed TZDs and L-tyrosine based agonsists. In the case of PPARα, PPARα does not bind ligands with large carboxylate head groups because of <scene name='Peroxisome_Proliferator-Activated_Receptors/Tyr_314/3'> Tyrosine 314</scene> as compared to PPARγ which has a smaller equivalent residue in His323.<ref>PMID:17317294</ref> Or in the case of binding some benzenesulfonamide derivatives, the <scene name='Peroxisome_Proliferator-Activated_Receptors/Pi_stacking/2'>pi stacking of Phe363 and the aromatic moiety</scene> in the case of PPARγ is lost in PPARα (Ile354) and PPARδ(Ile 363)<ref>PMID:16640330</ref> | Despite over 80% of the ligand binding cavity residues being conserved over all PPAR isotypes, it is the remaining 20% that creates the ligand specificity seen between isotypes. A few examples illustrate this point. In PPARδ, the cavity is significantly narrower adjacent to the AF-2 helix and Arm I. This prevents PPARδ from accommodating large headed TZDs and L-tyrosine based agonsists. In the case of PPARα, PPARα does not bind ligands with large carboxylate head groups because of <scene name='Peroxisome_Proliferator-Activated_Receptors/Tyr_314/3'> Tyrosine 314</scene> as compared to PPARγ which has a smaller equivalent residue in His323.<ref>PMID:17317294</ref> Or in the case of binding some benzenesulfonamide derivatives, the <scene name='Peroxisome_Proliferator-Activated_Receptors/Pi_stacking/2'>pi stacking of Phe363 and the aromatic moiety</scene> in the case of PPARγ is lost in PPARα (Ile354) and PPARδ(Ile 363)<ref>PMID:16640330</ref> | ||
Revision as of 09:55, 12 August 2010
Template:STRUCTURE 3dzy
The Peroxisome Proliferator-Activated Receptors (PPAR) α, δ, and γ are members of the nuclear receptor family. Since their discovery in the early 90s, it has become clear that the PPARs are essential modulators of environmental and dietary stimuli, acting as transcription factor to regulate mammalian metabolism, cellular differentiation, and tumorigenesis. The PPARs are the targets of numerous pharmaceutical drugs aimed at treating hypolipidemia and diabetes among other diseases.[1]
Contents |
Biological Role
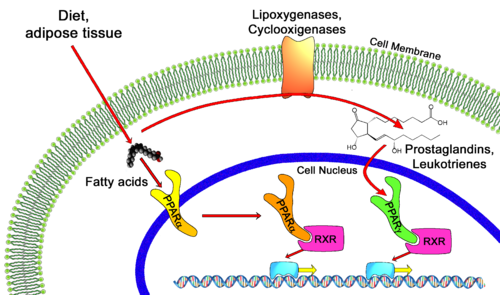
Transcription of individual genes in eukaryotic cells is controlled very precisely at a number of different levels. One key level is the binding of specific DNA binding transcriptional factors such as nuclear receptors, to facilitate RNA polymerase function. Unliganded PPARs form a heterodimer with retinoid X receptor (RXR), specifically RXRα, and bind to the Peroxisome Proliferator Response Element (PPRE), a specific DNA sequence present in the promoter region of PPAR-regulated genes, repressing transcription. [2] Also associated with this unliganded heterodimer is a co-repressor complex which possesses histone deacetylation activity, enforcing a tight chromatin structure which prevents gene transcription. [3] This co-repressor complex is released upon ligand binding (typical ligands include lipids and eicosanoids), allowing various co-activators and co-activator-associated proteins to be recruited to the scene. These protein complexes modulate chromatin remodeling and facilitate DNA unwinding and linkage to RNA polymerase II machinery to commence transcription. Some PPAR related co-activators include CBP (Histone Acetylation), SRC-1,2,3 (Chromatin Acetylation), [4], PGC-1 (Recruit HAT activities), PRIC285,320 (Chromatin Remodeling via Helicase activity)[5]and PIMT (RNA Capping via methyltransferase activity)[6].
PPARs regulate diverse biological processes varying from lipid and carbohydrate metabolism to inflammation and wound healing. While PPARα is the major regulator of fatty acid oxidation and uptake in the liver, PPARγ is expressed at extremely high levels in adipose tissue, macrophages, and the large intestine and controls lipid adipogenesis and energy conversion. PPARδ is expressed in most tissues and plays diverse roles involved in metabolism and wound healing. These nuclear receptors are of critical importance to the body as exemplified by PPARα knockdown mice suffering from a variety of metabolic defects including hypothermia, elevated plasma free fatty acid levels, and hypoglycemia, ultimately leading to death.[7]
Natural Ligands
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
David Canner, Michal Harel, Alexander Berchansky, Joel L. Sussman