Copper Amine Oxidase
From Proteopedia
| Line 8: | Line 8: | ||
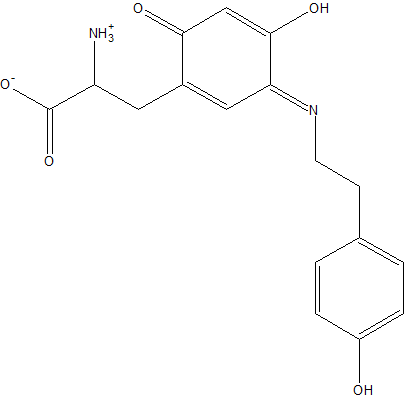
| - | [[Image:TTS.png|right|frame|alt=3-((3E)-4-HYDROXY-3-{[2-(4-HYDROXYPHENYL)ETHYL]IMINO}-6-OXOCYCLOHEXA-1,4-DIEN-1-YL)ALANINE.|Residue 382 is a modified tyrosine residue.]] 2d1w is a [http://en.wikipedia.org/wiki/Amine_oxidase_%28copper-containing%29 copper amine oxidase] derived from [http://en.wikipedia.org/wiki/Arthrobacter_globiformis Arthrobacter globiformis], and is classified as classified as EC 1.4.3.6 in the EC number classification of enzymes. It belongs to the larger class of [http://en.wikipedia.org/wiki/Oxidoreductase oxidoreductases]. The structure of this enzyme was determined by Murakawa et al. in 2005, by x-ray diffraction<ref>PMID:16487484</ref>. It consists of a disulfide-linked homodimer, with each subunit containing 638 residues. Each subunit also contains a copper ligand,<scene name='Sandbox_Reserved_331/Copper_ligand/5'> shown here</scene>, near the active site, which is coordinated by three histidine residues. Located near the Cu<sup>2+</sup> ligand is a tyrosine residue that has been modified into topa quinone, which is also a cofactor in all other copper amine oxidases<ref>PMID:8591028</ref>. The active site of the enzyme is located near the center of the homodimer, which is connected to the outside of the enzyme by an extensively hydrated channel. It is suspected that the water helps to carry O<sub>2</sub> to the active site, as well as being used in the reaction itself<ref>PMID:12135347</ref>. | + | [[Image:TTS.png|right|frame|alt=3-((3E)-4-HYDROXY-3-{[2-(4-HYDROXYPHENYL)ETHYL]IMINO}-6-OXOCYCLOHEXA-1,4-DIEN-1-YL)ALANINE.|Residue 382 is a modified tyrosine residue.]] 2d1w is a [http://en.wikipedia.org/wiki/Amine_oxidase_%28copper-containing%29 copper amine oxidase] derived from [http://en.wikipedia.org/wiki/Arthrobacter_globiformis Arthrobacter globiformis], and is classified as classified as EC 1.4.3.6 in the EC number classification of enzymes. It belongs to the larger class of [http://en.wikipedia.org/wiki/Oxidoreductase oxidoreductases]. The structure of this enzyme was determined by Murakawa et al. in 2005, by x-ray diffraction<ref>PMID:16487484</ref>. It consists of a disulfide-linked homodimer, with each subunit containing 638 residues. Each subunit also contains a copper ligand,<scene name='Sandbox_Reserved_331/Copper_ligand/5'> shown here</scene>, near the active site, which is coordinated by three histidine residues. Located near the Cu<sup>2+</sup> ligand is a tyrosine residue that has been modified into topa-quinone, which is also a cofactor in all other copper amine oxidases<ref>PMID:8591028</ref>. The active site of the enzyme is located near the center of the homodimer, which is connected to the outside of the enzyme by an extensively hydrated channel. It is suspected that the water helps to carry O<sub>2</sub> to the active site, as well as being used in the reaction itself<ref>PMID:12135347</ref>. |
== Reaction == | == Reaction == | ||
| Line 18: | Line 18: | ||
=== Mechanism === | === Mechanism === | ||
==== Reductive half-reaction ==== | ==== Reductive half-reaction ==== | ||
| - | In the reductive half-reaction, the carbonyl of topa quinone reacts with the primary amine of the substrate, forming the substrate Schiff-base intermediate. The α-proton is then abstracted by a conserved aspartic acid residue (Asp298), which forms the product Schiff-base intermediate. Several studies have shown that the abstraction of the α-proton proceeds through [http://en.wikipedia.org/wiki/Quantum_tunnelling quantum-mechanical tunneling], which allows the proton to pass through an energy barrier, which it would otherwise not be able to pass according to classical mechanics<ref>PMID:16487484</ref><ref>PMID:2790014</ref>.This bond is then hydrolyzed, which yields the aldehyde form of the substrate, | + | In the reductive half-reaction, the carbonyl of topa-quinone reacts with the primary amine of the substrate, forming the substrate Schiff-base intermediate. The α-proton is then abstracted by a conserved aspartic acid residue (Asp298), which forms the product Schiff-base intermediate. Several studies have shown that the abstraction of the α-proton proceeds through [http://en.wikipedia.org/wiki/Quantum_tunnelling quantum-mechanical tunneling], which allows the proton to pass through an energy barrier, which it would otherwise not be able to pass according to classical mechanics<ref>PMID:16487484</ref><ref>PMID:2790014</ref>.This bond is then hydrolyzed, which yields the aldehyde form of the substrate, the aminoquinol form of topa-quinone, and a proton. |
==== Oxidative half-reaction ==== | ==== Oxidative half-reaction ==== | ||
| - | + | In the oxidative half-reaction, electrons are transferred from the reduced form of topa-quinone to O<sub>2</sub>, forming a superoxide ion that is stabilized by the Cu<sup>2+</sup> ligand. Reduction of the superoxide by the amino-quinone form of topa-quinone yields peroxide and the imino-quinone form, leading to the release of hydrogen peroxide. Hydrolysis of the imino-quinone form of topa-quinone then releases free ammonia, and regenerates the oxidized form of topa-quinone<ref>PMID:2790014</ref>. | |
== Additional Resources == | == Additional Resources == | ||
*[http://www.pdb.org/pdb/explore/explore.do?structureId=2D1W Entry 2D1W in the RCSB Protein Data Bank] | *[http://www.pdb.org/pdb/explore/explore.do?structureId=2D1W Entry 2D1W in the RCSB Protein Data Bank] | ||
Revision as of 22:00, 31 March 2011
| This Sandbox is Reserved from January 10, 2010, through April 10, 2011 for use in BCMB 307-Proteins course taught by Andrea Gorrell at the University of Northern British Columbia, Prince George, BC, Canada. |
To get started:
More help: Help:Editing |
Contents |
Structure
2d1w is a copper amine oxidase derived from Arthrobacter globiformis, and is classified as classified as EC 1.4.3.6 in the EC number classification of enzymes. It belongs to the larger class of oxidoreductases. The structure of this enzyme was determined by Murakawa et al. in 2005, by x-ray diffraction[1]. It consists of a disulfide-linked homodimer, with each subunit containing 638 residues. Each subunit also contains a copper ligand,, near the active site, which is coordinated by three histidine residues. Located near the Cu2+ ligand is a tyrosine residue that has been modified into topa-quinone, which is also a cofactor in all other copper amine oxidases[2]. The active site of the enzyme is located near the center of the homodimer, which is connected to the outside of the enzyme by an extensively hydrated channel. It is suspected that the water helps to carry O2 to the active site, as well as being used in the reaction itself[3].Reaction
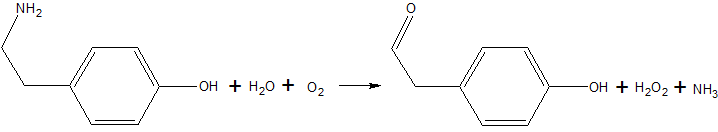
Overview
Copper amine oxidase catalyzes the oxidation of a primary amine to the corresponding aldehyde, yielding hydrogen peroxide and free ammonia. An example of this is the oxidation of tyramine:
Mechanism
Reductive half-reaction
In the reductive half-reaction, the carbonyl of topa-quinone reacts with the primary amine of the substrate, forming the substrate Schiff-base intermediate. The α-proton is then abstracted by a conserved aspartic acid residue (Asp298), which forms the product Schiff-base intermediate. Several studies have shown that the abstraction of the α-proton proceeds through quantum-mechanical tunneling, which allows the proton to pass through an energy barrier, which it would otherwise not be able to pass according to classical mechanics[4][5].This bond is then hydrolyzed, which yields the aldehyde form of the substrate, the aminoquinol form of topa-quinone, and a proton.
Oxidative half-reaction
In the oxidative half-reaction, electrons are transferred from the reduced form of topa-quinone to O2, forming a superoxide ion that is stabilized by the Cu2+ ligand. Reduction of the superoxide by the amino-quinone form of topa-quinone yields peroxide and the imino-quinone form, leading to the release of hydrogen peroxide. Hydrolysis of the imino-quinone form of topa-quinone then releases free ammonia, and regenerates the oxidized form of topa-quinone[6].
Additional Resources
- Entry 2D1W in the RCSB Protein Data Bank
- The Biosynthesis of Topa Quinone Cofactor in Bacterial Amine Oxidases
References
- ↑ Murakawa T, Okajima T, Kuroda S, Nakamoto T, Taki M, Yamamoto Y, Hayashi H, Tanizawa K. Quantum mechanical hydrogen tunneling in bacterial copper amine oxidase reaction. Biochem Biophys Res Commun. 2006 Apr 7;342(2):414-23. Epub 2006 Feb 8. PMID:16487484 doi:10.1016/j.bbrc.2006.01.150
- ↑ Parsons MR, Convery MA, Wilmot CM, Yadav KD, Blakeley V, Corner AS, Phillips SE, McPherson MJ, Knowles PF. Crystal structure of a quinoenzyme: copper amine oxidase of Escherichia coli at 2 A resolution. Structure. 1995 Nov 15;3(11):1171-84. PMID:8591028
- ↑ Mure M, Mills SA, Klinman JP. Catalytic mechanism of the topa quinone containing copper amine oxidases. Biochemistry. 2002 Jul 30;41(30):9269-78. PMID:12135347
- ↑ Murakawa T, Okajima T, Kuroda S, Nakamoto T, Taki M, Yamamoto Y, Hayashi H, Tanizawa K. Quantum mechanical hydrogen tunneling in bacterial copper amine oxidase reaction. Biochem Biophys Res Commun. 2006 Apr 7;342(2):414-23. Epub 2006 Feb 8. PMID:16487484 doi:10.1016/j.bbrc.2006.01.150
- ↑ Grant KL, Klinman JP. Evidence that both protium and deuterium undergo significant tunneling in the reaction catalyzed by bovine serum amine oxidase. Biochemistry. 1989 Aug 8;28(16):6597-605. PMID:2790014
- ↑ Grant KL, Klinman JP. Evidence that both protium and deuterium undergo significant tunneling in the reaction catalyzed by bovine serum amine oxidase. Biochemistry. 1989 Aug 8;28(16):6597-605. PMID:2790014
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Raymond Lyle, Alexander Berchansky, OCA, Jaime Prilusky