Glyceraldehyde-3-Phosphate Dehydrogenase
From Proteopedia
| Line 20: | Line 20: | ||
GAP + NAD+ + Pi +GAPDH <==> 1,3-bisphosphoglycerate + NADH | GAP + NAD+ + Pi +GAPDH <==> 1,3-bisphosphoglycerate + NADH | ||
| - | + | == Reaction Mechanism == | |
The mechanism of the glycolysis reaction is fairly straight forward. After the aldehyde enters the <scene name='Nathan_Line_sandbox_3/Active_site/3'>active site</scene> (highlighted in green), the sulfhydryl group from <scene name='Nathan_Line_sandbox_3/Cystine/7'>Cystine 151</scene> attacks the nucleophilic carbon to form a thiohemiacetal. This intermediate undergoes oxidation due to a hydride transfer to a nearby NAD+ forming a thioester. From here, a phosphate group enters and attacks the same carbonyl while at the same time it is separated from the cystine by the protonated <scene name='Nathan_Line_sandbox_3/Histidine/5'>Histidine 178</scene> group. This produces the desired 1,3-bisphosphoglycerate. Though cystine-151 and histidine-178 are direct contributers to the catalytic process, other residues also influence the activity of this enzyme indirectly. <scene name='Nathan_Line_sandbox_3/Other/2'>Thr-210 and Arg-233</scene> are two such residues that contribute to the binding of the reactants rather than the catalytic mechanism. Regulation of GAPDH occurs through its coupling with the PGK reaction. This coupling is needed due to the slightly positive delta G of the glycolysis. The larger negative delta G of the PGK reaction results in the following overall net reaction with a delta G of -12.1 kJ/mol: | The mechanism of the glycolysis reaction is fairly straight forward. After the aldehyde enters the <scene name='Nathan_Line_sandbox_3/Active_site/3'>active site</scene> (highlighted in green), the sulfhydryl group from <scene name='Nathan_Line_sandbox_3/Cystine/7'>Cystine 151</scene> attacks the nucleophilic carbon to form a thiohemiacetal. This intermediate undergoes oxidation due to a hydride transfer to a nearby NAD+ forming a thioester. From here, a phosphate group enters and attacks the same carbonyl while at the same time it is separated from the cystine by the protonated <scene name='Nathan_Line_sandbox_3/Histidine/5'>Histidine 178</scene> group. This produces the desired 1,3-bisphosphoglycerate. Though cystine-151 and histidine-178 are direct contributers to the catalytic process, other residues also influence the activity of this enzyme indirectly. <scene name='Nathan_Line_sandbox_3/Other/2'>Thr-210 and Arg-233</scene> are two such residues that contribute to the binding of the reactants rather than the catalytic mechanism. Regulation of GAPDH occurs through its coupling with the PGK reaction. This coupling is needed due to the slightly positive delta G of the glycolysis. The larger negative delta G of the PGK reaction results in the following overall net reaction with a delta G of -12.1 kJ/mol: | ||
Revision as of 14:51, 19 April 2011
| |||||||||
| 3gpd, resolution 3.50Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Activity: | Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating), with EC number 1.2.1.12 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Function
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a very important enzyme in the production of energy. This enzyme catalyzes the sixth step in the process of breaking down glucose into energy, also known as glycolysis. Though this is its main function, GAPDH has been shown to perform other functions including transcription activation, initiation of apoptosis, and ER to Golgi apparatus vesicle transportation. However, this page will focus on GAPDH’s role in glycolysis.
Structure
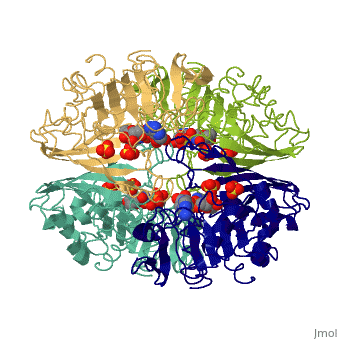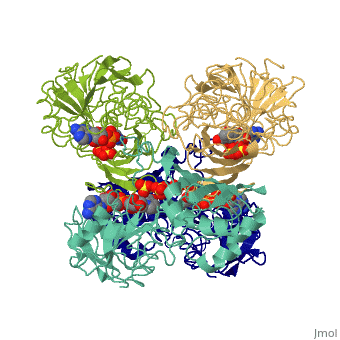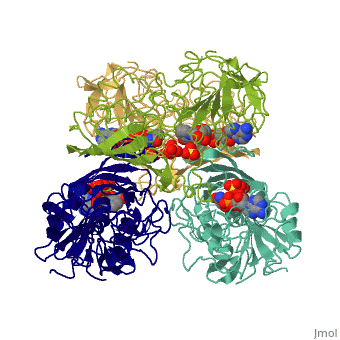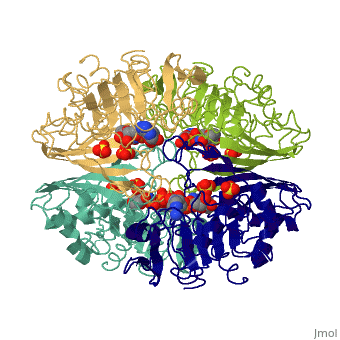
GAPDH most commonly exists as what looks to be a dimer. Interesting though, the two monomers of the enzyme are not exactly the same. While one side consists only of parallel and antiparallel beta-sheets, the other monomer is made up of both . Though each monomer does not have to exact same sequence, each does contain replicate active sites and function. This is consistent with the following SCOP information:
Class: Alpha and beta proteins (a/b) Fold: NAD(P)-binding Rossmann-fold domains Superfamily: NAD(P)-binding Rossmann-fold domains Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminus domain Protein: Glyceraldehyde-3-phosphate dehydrogenase Species: Human
The specific reaction that GAPDH catalyzes is shown below:
GAP + NAD+ + Pi +GAPDH <==> 1,3-bisphosphoglycerate + NADH
Reaction Mechanism
The mechanism of the glycolysis reaction is fairly straight forward. After the aldehyde enters the (highlighted in green), the sulfhydryl group from attacks the nucleophilic carbon to form a thiohemiacetal. This intermediate undergoes oxidation due to a hydride transfer to a nearby NAD+ forming a thioester. From here, a phosphate group enters and attacks the same carbonyl while at the same time it is separated from the cystine by the protonated group. This produces the desired 1,3-bisphosphoglycerate. Though cystine-151 and histidine-178 are direct contributers to the catalytic process, other residues also influence the activity of this enzyme indirectly. are two such residues that contribute to the binding of the reactants rather than the catalytic mechanism. Regulation of GAPDH occurs through its coupling with the PGK reaction. This coupling is needed due to the slightly positive delta G of the glycolysis. The larger negative delta G of the PGK reaction results in the following overall net reaction with a delta G of -12.1 kJ/mol:
GAP + Pi + NAD+ + ADP ==> 3PG + NADH + ATP
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
1) Voet, D, Voet, J, & Pratt, C. (2008). Fundamentals of biochemistry, third edition. Hoboken, NJ: Wiley & Sons, Inc.
2)Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain. Retrived from: http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.d.c.b.d.html
3)- Kundu S, Roy D. Computational study of glyceraldehyde-3-phosphate dehydrogenase of Entamoeba histolytica: implications for structure-based drug design. J Biomol Struct Dyn. 2007 Aug;25(1):25-33. PMID:17676935
- Vospelnikova ND, Safronova MI, Shuvalova ER, Baratova LA, Kniazev SP, Nagradova NK. Identification of an arginine residue important for catalytic activity in the primary structure of D-glyceraldehyde 3-phosphate dehydrogenase. Studies with the rat skeletal-muscle enzyme. Biochem J. 1981 Dec 1;199(3):757-65. PMID:7340828
- Butterfield DA, Hardas SS, Lange ML. Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer's disease: many pathways to neurodegeneration. J Alzheimers Dis. 2010;20(2):369-93. PMID:20164570 doi:10.3233/JAD-2010-1375
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Nathan Line, Andrew Swart, Alexander Berchansky, Alice Harmon, David Canner