Taylor Gal4 Sandbox
From Proteopedia
(→DNA RECOGNITION BY GAL4: STRUCTURE OF A PROTEIN/DNA COMPLEX) |
|||
| (2 intermediate revisions not shown.) | |||
| Line 11: | Line 11: | ||
===DNA RECOGNITION BY GAL4: STRUCTURE OF A PROTEIN/DNA COMPLEX=== | ===DNA RECOGNITION BY GAL4: STRUCTURE OF A PROTEIN/DNA COMPLEX=== | ||
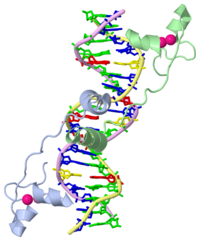
| - | GAL4 is a transcription factor that induces genes required for the metabolism of galactose, specifically enzymes involved in the conversion of galactose to glucose. This structure is for the DNA binding domain of GAL4, and contains 65 residues from the N terminus. The protein binds as a <scene name='Taylor_Gal4_Sandbox/Dimer/1'>dimer</scene> to a symmetrical 17-base-pair sequence. | + | GAL4 is a transcription factor that induces genes required for the metabolism of galactose, specifically enzymes involved in the conversion of galactose to glucose. This structure is for the DNA binding domain of GAL4, and contains 65 residues from the N terminus. The protein binds as a <scene name='Taylor_Gal4_Sandbox/Dimer/1'>dimer</scene> to a symmetrical 17-base-pair sequence. Each subunit folds into three distinct modules: a compact, <scene name='Taylor_Gal4_Sandbox/Metal_binding_domain/2'>metal binding domain</scene>(residues 8-40), an extended <scene name='Taylor_Gal4_Sandbox/Linker/1'>linker</scene>(41-49), and an <scene name='Taylor_Gal4_Sandbox/Dimerization/1'>alpha-helical dimerization element</scene> (50-64). The small, <scene name='Taylor_Gal4_Sandbox/Zn_binding/1'>Zn(2+)-containing domain</scene>, which contains two metal ions tetrahedrally coordinated by six cysteines. This metal binding domain recognizes a conserved CCG triplet at each end of the site through direct contacts with the <scene name='Taylor_Gal4_Sandbox/Major_groove/1'>major groove</scene>. A short coiled-coil dimerization element imposes 2-fold symmetry. A segment of extended polypeptide chain links the metal-binding module to the dimerization element and specifies the length of the site. The relatively open structure of the complex would allow another protein to bind coordinately with GAL4. |
| - | + | ||
==About this Structure== | ==About this Structure== | ||
Current revision
| |||||||||
| 1d66, resolution 2.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
DNA RECOGNITION BY GAL4: STRUCTURE OF A PROTEIN/DNA COMPLEX
GAL4 is a transcription factor that induces genes required for the metabolism of galactose, specifically enzymes involved in the conversion of galactose to glucose. This structure is for the DNA binding domain of GAL4, and contains 65 residues from the N terminus. The protein binds as a to a symmetrical 17-base-pair sequence. Each subunit folds into three distinct modules: a compact, (residues 8-40), an extended (41-49), and an (50-64). The small, , which contains two metal ions tetrahedrally coordinated by six cysteines. This metal binding domain recognizes a conserved CCG triplet at each end of the site through direct contacts with the . A short coiled-coil dimerization element imposes 2-fold symmetry. A segment of extended polypeptide chain links the metal-binding module to the dimerization element and specifies the length of the site. The relatively open structure of the complex would allow another protein to bind coordinately with GAL4.
About this Structure
1d66 is a 4 chain structure with sequence from Saccharomyces cerevisiae. Full crystallographic information is available from OCA.
See Also
Reference
- Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992 Apr 2;356(6368):408-14. PMID:1557122 doi:http://dx.doi.org/10.1038/356408a0