AChE inhibitors and substrates
From Proteopedia
| Line 13: | Line 13: | ||
in 1991 <ref name="Sussman">PMID:1678899</ref> opened up new horizons in research on an [http://en.wikipedia.org/wiki/Enzyme enzyme] that had already been the subject of intensive investigation. The unanticipated structure of this extremely rapid enzyme, in which the [http://en.wikipedia.org/wiki/Active_site active site] was found to be buried at the bottom of a <scene name='2ace/Active_site/3'>deep and narrow gorge</scene>, lined by <scene name='2ace/Active_site/4'>14 aromatic residues</scene> <font color='darkmagenta'><b>(colored dark magenta)</b></font>, led to a revision of the views then held concerning [http://en.wikipedia.org/wiki/Substrate_(biochemistry) substrate] traffic, recognition and hydrolysis <ref name="Botti">PMID:10545346</ref>. This led to a series of theoretical and experimental studies, which took advantage of recent advances in theoretical techniques for treatment of [http://en.wikipedia.org/wiki/Protein proteins], such as | in 1991 <ref name="Sussman">PMID:1678899</ref> opened up new horizons in research on an [http://en.wikipedia.org/wiki/Enzyme enzyme] that had already been the subject of intensive investigation. The unanticipated structure of this extremely rapid enzyme, in which the [http://en.wikipedia.org/wiki/Active_site active site] was found to be buried at the bottom of a <scene name='2ace/Active_site/3'>deep and narrow gorge</scene>, lined by <scene name='2ace/Active_site/4'>14 aromatic residues</scene> <font color='darkmagenta'><b>(colored dark magenta)</b></font>, led to a revision of the views then held concerning [http://en.wikipedia.org/wiki/Substrate_(biochemistry) substrate] traffic, recognition and hydrolysis <ref name="Botti">PMID:10545346</ref>. This led to a series of theoretical and experimental studies, which took advantage of recent advances in theoretical techniques for treatment of [http://en.wikipedia.org/wiki/Protein proteins], such as | ||
[http://en.wikipedia.org/wiki/Molecular_dynamics molecular dynamics] and [http://en.wikipedia.org/wiki/Electrostatics electrostatics] and to [http://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis], utilizing suitable expression | [http://en.wikipedia.org/wiki/Molecular_dynamics molecular dynamics] and [http://en.wikipedia.org/wiki/Electrostatics electrostatics] and to [http://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis], utilizing suitable expression | ||
| - | systems. | + | systems. [http://en.wikipedia.org/wiki/Acetylcholinesterase Acetylcholinesterase] [http://en.wikipedia.org/wiki/Hydrolysis hydrolysizes] the [http://en.wikipedia.org/wiki/Neurotransmitter neurotransmitter] [http://en.wikipedia.org/wiki/Acetylcholine acetylcholine] <scene name='2ace/Cv/2'>(ACh)</scene>, producing <scene name='2ace/Cv/3'>choline and an acetate</scene> group. ACh directly binds <scene name='2ace/Cv/4'>Ser200</scene> (via its [http://en.wikipedia.org/wiki/Nucleophile nucleophilic] Oγ atom) within the <scene name='2ace/Cv/5'>catalytic triad (Ser200, His440, and Glu327)</scene> (ACh/''Tc''AChE structure [[2ace]]). The residues <scene name='2ace/Cv/6'>Trp84 and Phe330</scene> are also important in the [http://en.wikipedia.org/wiki/Ligand ligand] recognition <ref name="Raves">PMID:8989325</ref>. After this binding acetylcholinesterase <scene name='2ace/Cv/7'>hydrolysizes</scene> ACh.{{Clear}} |
==AChE monovalent inhibitors== | ==AChE monovalent inhibitors== | ||
Revision as of 09:48, 27 October 2011

Dear readers, this page presents only a small part of the great world of the acetylcholinesterase inhibitors. So, please see also our pages AChE inhibitors and substrates (Part II), AChE inhibitors and substrates (Part III), AChE bivalent inhibitors and AChE bivalent inhibitors (Part II). The images at the left and at the right correspond to one representative acetylcholinesterase with substrate structure, i.e. crystal structure of Torpedo californica acetylcholinesterase with acetylcholine (2ace).
Contents |
AChE substrate
|
Solution of the three-dimensional (3D) structure of Torpedo californica acetylcholinesterase (TcAChE) in 1991 [1] opened up new horizons in research on an enzyme that had already been the subject of intensive investigation. The unanticipated structure of this extremely rapid enzyme, in which the active site was found to be buried at the bottom of a , lined by (colored dark magenta), led to a revision of the views then held concerning substrate traffic, recognition and hydrolysis [2]. This led to a series of theoretical and experimental studies, which took advantage of recent advances in theoretical techniques for treatment of proteins, such as molecular dynamics and electrostatics and to site-directed mutagenesis, utilizing suitable expression
systems. Acetylcholinesterase hydrolysizes the neurotransmitter acetylcholine , producing group. ACh directly binds (via its nucleophilic Oγ atom) within the (ACh/TcAChE structure 2ace). The residues are also important in the ligand recognition [3]. After this binding acetylcholinesterase ACh.AChE monovalent inhibitors
Organophosphorus acid anhydride nerve agents
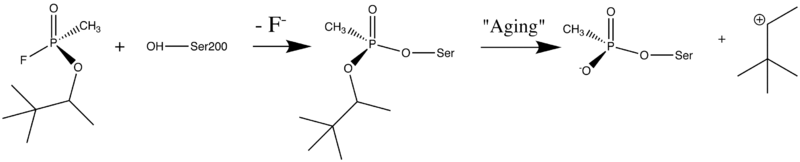
Organophosphorus (OP) acid anhydride nerve agents are potent inhibitors which rapidly phosphonylate AChE and then may undergo an internal dealkylation reaction (called "aging") to produce an OP-enzyme conjugate that cannot be reactivated.
| |||||||||||
Treatment of Alzheimer's disease
Please see pages AChE inhibitors and substrates (Part II) and AChE inhibitors and substrates (Part III).
AChE bivalent inhibitors
Please see pages AChE bivalent inhibitors and AChE bivalent inhibitors (Part II)
Selected 3D Structures of AChE
- 2ace This is the original solved structure for Torpedo Californica
- 1ea5 This is one of the highest quality representative X-ray structures in the PDB.
- 1eve The E2020 (Aricept) complex.
- 1ax9 Endrophonium complex.
- 1vot Complex with Huperzine, a Chinese folk medicine.
- 1fss Complex with snake venum toxin Fasciculin-II.
- 1acj Complex with tacrine.
- 1e66 Complex with huprine X.
- 1dx6 Complex with galanthamine.
- 1qti Complex with galanthamine.
- 1w6r Complex with galanthamine iminium derivative.
- 2ack Complex with edrophonium.
- 1vzj Structure of the tetramerization domain of acetylcholinesterase.
- 1gqr Complex with rivastigmine.
- 1gqs Complex with NAP alone.
- 1vxr Complex with VX.
- 2vja Complex with OTMA.
- 1som Complex with soman.
- 2wfz Complex with nonaged soman.
- 2wg0 Complex with aged soman.
- 2wg1 Complex with aged soman and 2-PAM.
- 1cfj Complex with aged sarin.
- 2dfp Complex with aged DFP.
Additional Resources
For additional information, see: Alzheimer's Disease
References
- ↑ Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872-9. PMID:1678899
- ↑ Botti SA, Felder CE, Lifson S, Sussman JL, Silman I. A modular treatment of molecular traffic through the active site of cholinesterase. Biophys J. 1999 Nov;77(5):2430-50. PMID:10545346
- ↑ Raves ML, Harel M, Pang YP, Silman I, Kozikowski AP, Sussman JL. Structure of acetylcholinesterase complexed with the nootropic alkaloid, (-)-huperzine A. Nat Struct Biol. 1997 Jan;4(1):57-63. PMID:8989325
- ↑ Sanson B, Nachon F, Colletier JP, Froment MT, Toker L, Greenblatt HM, Sussman JL, Ashani Y, Masson P, Silman I, Weik M. Crystallographic Snapshots of Nonaged and Aged Conjugates of Soman with Acetylcholinesterase, and of a Ternary Complex of the Aged Conjugate with Pralidoxime (dagger). J Med Chem. 2009 Jul 30. PMID:19642642 doi:10.1021/jm900433t
- ↑ 5.0 5.1 5.2 Millard CB, Kryger G, Ordentlich A, Greenblatt HM, Harel M, Raves ML, Segall Y, Barak D, Shafferman A, Silman I, Sussman JL. Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level. Biochemistry. 1999 Jun 1;38(22):7032-9. PMID:10353814 doi:http://dx.doi.org/10.1021/bi982678l
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Joel L. Sussman, Michal Harel, Jaime Prilusky, David Canner