Journal:PLoS ONE:2
From Proteopedia
(Difference between revisions)

| Line 11: | Line 11: | ||
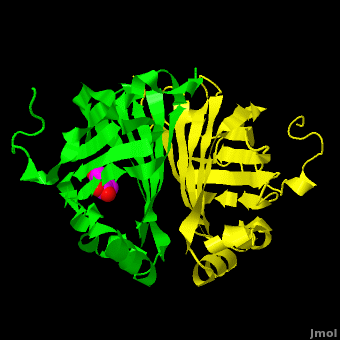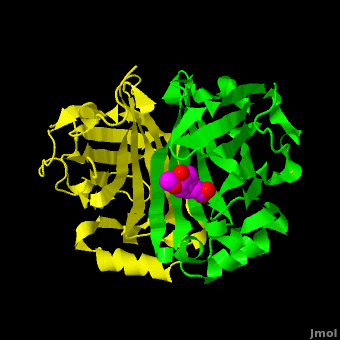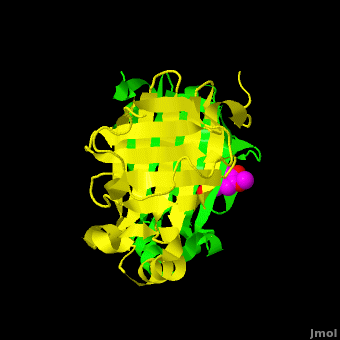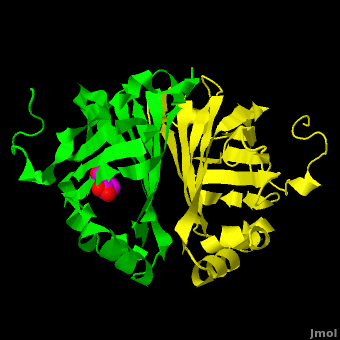
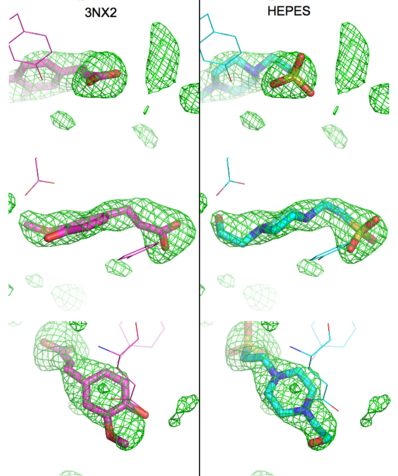
The editors of the journal were contacted by a reader, who raised concerns about the accuracy of the assignment of the density in the <scene name='Journal:PLoS_ONE:2/Cv1/6'>active site of chain A</scene> of [[3nx2]]. The reader suggested that the density was more reminiscent of a <scene name='Journal:PLoS_ONE:2/Cv1/8'>HEPES molecule</scene> (used in the crystallization buffer) based on the shape of the density, and the fact that the stereochemistry in the region of the double bond (between atoms C7 and C8) was distorted. <font color='red'><b>O atoms are colored in red</b></font>, <font color='blue'><b>N atoms are in blue</b></font>, <span style="color:yellow;background-color:black;font-weight:bold;">S atom in yellow</span>; <span style="color:cyan;background-color:black;font-weight:bold;">C atoms of HEPES in cyan</span>, and <font color='magenta'><b>C atoms of Ferulate in magenta</b></font>. <scene name='Journal:PLoS_ONE:2/Cv1/10'>To see possible superposition of HEPES on Ferulate click here</scene>. In response to these concerns the Editors contacted one of us (JLS), asking for a critical review of the claims of the authors regarding [[3nx2]]. The results of this review are detailed below. | The editors of the journal were contacted by a reader, who raised concerns about the accuracy of the assignment of the density in the <scene name='Journal:PLoS_ONE:2/Cv1/6'>active site of chain A</scene> of [[3nx2]]. The reader suggested that the density was more reminiscent of a <scene name='Journal:PLoS_ONE:2/Cv1/8'>HEPES molecule</scene> (used in the crystallization buffer) based on the shape of the density, and the fact that the stereochemistry in the region of the double bond (between atoms C7 and C8) was distorted. <font color='red'><b>O atoms are colored in red</b></font>, <font color='blue'><b>N atoms are in blue</b></font>, <span style="color:yellow;background-color:black;font-weight:bold;">S atom in yellow</span>; <span style="color:cyan;background-color:black;font-weight:bold;">C atoms of HEPES in cyan</span>, and <font color='magenta'><b>C atoms of Ferulate in magenta</b></font>. <scene name='Journal:PLoS_ONE:2/Cv1/10'>To see possible superposition of HEPES on Ferulate click here</scene>. In response to these concerns the Editors contacted one of us (JLS), asking for a critical review of the claims of the authors regarding [[3nx2]]. The results of this review are detailed below. | ||
| - | The structure of [[3nx2]] was downloaded from the PDB as well as the structure factors. The software suite PHENIX<ref name="Adams">PMID:20124702</ref> was used to run simulated annealing on the structure, with the ligand removed. This produced a simulated annealing Fo - Fc omit map, which should have all structural bias caused by the original ligand removed. A HEPES molecule was fitted into this density using COOT<ref name="Emsley">PMID:20383002</ref>, and refined in PHENIX (R = 19.4%, Rfree = 24.4%). Although the R and Rfree are slightly higher than for [[3nx2]] (R =18.9%, Rfree = 23.6%), this is to be expected, since the geometry of the model refined in PHENIX (rms bonds 0.007, angles 1.12) was much tighter than the one obtained from REFMAC<ref name="Murshudov">PMID:15299926</ref> (rms bonds 0.019, angles 1.81). In Figure (see below), we superimpose the omit map with the two refined structures: on the left, [[3nx2]], with magenta carbon atoms, and on the right, with cyan carbon atoms, the PHENIX-refined enzyme-HEPES complex. <scene name='Journal:PLoS_ONE:2/Cv5/ | + | The structure of [[3nx2]] was downloaded from the PDB as well as the structure factors. The software suite PHENIX<ref name="Adams">PMID:20124702</ref> was used to run simulated annealing on the structure, with the ligand removed. This produced a simulated annealing Fo - Fc omit map, which should have all structural bias caused by the original ligand removed. A HEPES molecule was fitted into this density using COOT<ref name="Emsley">PMID:20383002</ref>, and refined in PHENIX (R = 19.4%, Rfree = 24.4%). Although the R and Rfree are slightly higher than for [[3nx2]] (R =18.9%, Rfree = 23.6%), this is to be expected, since the geometry of the model refined in PHENIX (rms bonds 0.007, angles 1.12) was much tighter than the one obtained from REFMAC<ref name="Murshudov">PMID:15299926</ref> (rms bonds 0.019, angles 1.81). In Figure (see below), we superimpose the omit map with the two refined structures: on the left, [[3nx2]], with magenta carbon atoms, and on the right, with cyan carbon atoms, the PHENIX-refined enzyme-HEPES complex. <scene name='Journal:PLoS_ONE:2/Cv5/3'>See also 3D visualization of this superposition.</scene> |
{{Clear}} | {{Clear}} | ||
[[Image:Elden1.png|left|400px|thumb|'''Figure:''' Simulated annealing omit map (Fo – Fc) in region of active site, contoured at 3σ. On the left is the superposition of the structure of ferulate from [[3nx2]] (<font color='magenta'><b>magenta C atoms</b></font>), and on the right the refined HEPES molecule (<span style="color:cyan;background-color:black;font-weight:bold;">cyan C atoms</span>). The three views show different aspects of the density and modeled molecules. In addition we have included Tyr27 and Glu134 from each respective structure. Prepared with [http://www.pymol.org PyMOL].]] | [[Image:Elden1.png|left|400px|thumb|'''Figure:''' Simulated annealing omit map (Fo – Fc) in region of active site, contoured at 3σ. On the left is the superposition of the structure of ferulate from [[3nx2]] (<font color='magenta'><b>magenta C atoms</b></font>), and on the right the refined HEPES molecule (<span style="color:cyan;background-color:black;font-weight:bold;">cyan C atoms</span>). The three views show different aspects of the density and modeled molecules. In addition we have included Tyr27 and Glu134 from each respective structure. Prepared with [http://www.pymol.org PyMOL].]] | ||
Revision as of 11:59, 11 December 2011
| |||||||||||
- ↑ 1.0 1.1 1.2 Gu W, Yang J, Lou Z, Liang L, Sun Y, Huang J, Li X, Cao Y, Meng Z, Zhang KQ. Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4. PLoS One. 2011 Jan 21;6(1):e16262. PMID:21283705 doi:10.1371/journal.pone.0016262
- ↑ Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010 Feb;66(Pt 2):213-21. Epub 2010, Jan 22. PMID:20124702 doi:10.1107/S0907444909052925
- ↑ Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010 Apr;66(Pt 4):486-501. Epub 2010, Mar 24. PMID:20383002 doi:10.1107/S0907444910007493
- ↑ Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997 May 1;53(Pt 3):240-55. PMID:15299926 doi:http://dx.doi.org/10.1107/S0907444996012255
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.