This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Xuni Li/Sandbox 1
From Proteopedia
| (36 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <Structure load='2ch7' size='400' frame='true' align='right' caption='Cytoplasmic Domain of Thermotoga maritima receptor' scene='User:Xuni_Li/Sandbox_1/Initial/1' /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | <Structure load='2ch7' size=' | + | |
One of the [[CBI Molecules]] being studied in the [http://www.umass.edu/cbi/ University of Massachusetts Amherst Chemistry-Biology Interface Program] at UMass Amherst and on display at the [http://www.molecularplayground.org/ Molecular Playground]. | One of the [[CBI Molecules]] being studied in the [http://www.umass.edu/cbi/ University of Massachusetts Amherst Chemistry-Biology Interface Program] at UMass Amherst and on display at the [http://www.molecularplayground.org/ Molecular Playground]. | ||
| - | + | === Introduction === | |
| - | + | ---- | |
| - | + | Bacteria use their receptors to sense the environment to change their swimming patterns. There are different kinds of [http://en.wikipedia.org/wiki/Chemoreceptor chemoreceptors] that respond to different stimuli. The figure on the right is the [http://en.wikipedia.org/wiki/Methyl-accepting_chemotaxis_protein methyl-accepting protein] of Thermotoga maritima <scene name='User:Xuni_Li/Sandbox_1/Initial/1'>receptor</scene>. Bacteria like to flee away from the repellent when high concentrations are present in the environment. CheA is a [http://en.wikipedia.org/wiki/Histidine_kinase histidine kinase] that associates with CheW, an [http://en.wikipedia.org/wiki/Adaptor_protein adaptor protein], will cause the flagella to turn clockwise and result in a tumbling motion. On the other hand, when a high concentration of attractants are present in the environment, the CheA kinase will be turned off, cause flagella to turn counterclockwise, resulting in a forward swimming pattern. <ref name="introduction">Hazelbauer, Falke and Parkinson. "Bacterial chemoreceptors: high-performance signaling in networked arrays." Biochemical Sciences, 2007, 33 (1), 9-19. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/18165013]</ref> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | [[Image:Receptor.png]] | ||
| - | The | + | ===Structure and Functions=== |
| + | ---- | ||
| + | [http://en.wikipedia.org/wiki/Chemoreceptor Chemoreceptors] usually contain a periplasmic ligand binding domain, the transmembrane domain, a [http://www.ebi.ac.uk/interpro/IEntry?ac=IPR003660 HAMP domain] which is for signal conversion, and the cytoplasmic domain that contains the methylation sites, flexible bundle and protein binding sites where CheA and CheW bind. | ||
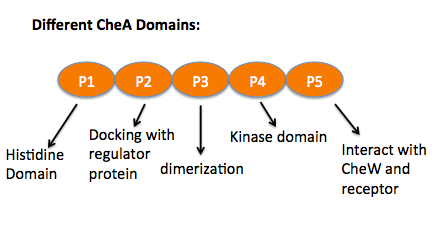
| + | CheA is a [http://en.wikipedia.org/wiki/Histidine_kinase histidine kinase] that contains five units. The P1 domain is the site of substrate [http://en.wikipedia.org/wiki/Autophosphorylation autophosphorylation] that associates with kinase P4 domain, P2 is where phosphate transfers to CheY, another response regulator protein, from P1. P3, P4 and P5 are the dimerization, kinase and the receptor-coupling domains. The P3 domain was predicted to interact with CheW which stabilize the interface between P3 and P5. The NMR structure has shown that P5 is proximal to the CheW β barrel (residues 635-660). <ref name="structure">Park, Borbat, Gonzalez-Bonet, Bhatnagar, et al. "Reconstruction of the chemotaxis receptor-kinase assembly." Nature Structural and Molecular Biology, April 23, 2006, 13 (5), 400-407. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/16622408]</ref> On the right, is the CheW with CheA P4, P5 domains (orange) <scene name='User:Xuni_Li/Sandbox_1/Initial2/1'>superimposed</scene> with CheA P3, P4, P5 (blue). | ||
| - | Molecular Playground | + | <scene name='User:Xuni_Li/Sandbox_1/Initial2/1'>Molecular Playground</scene> |
| + | Molecular Playground banner: P4, P5 of CheA binding with CheW superimpose with P3, P4 P5 of CheA | ||
| - | + | <Structure load='Xuni1.pdb' size='300' frame='true' align='right' caption='P4, P5 of CheA binding with CheW (orange) superimpose with P3, P4, P5 of CheA (blue)' scene='User:Xuni_Li/Sandbox_1/Initial2/1' /> | |
| - | < | + | |
| + | [[Image:CheA.png]] | ||
| - | === | + | === References === |
| + | ---- | ||
| + | <references/> | ||
| + | =Acknowledgement= | ||
| - | + | To Luis E Ramirez-Tapia his advice to develop this page. | |
| - | + | =See Also= | |
| + | *[[Molecular Playground/Bacterial Chemotaxis Receptors]] | ||
| + | *[[Molecular Playground/cytoplasmic domain of a serine chemotaxis receptor]] | ||
Current revision
|
One of the CBI Molecules being studied in the University of Massachusetts Amherst Chemistry-Biology Interface Program at UMass Amherst and on display at the Molecular Playground.
Contents |
Introduction
Bacteria use their receptors to sense the environment to change their swimming patterns. There are different kinds of chemoreceptors that respond to different stimuli. The figure on the right is the methyl-accepting protein of Thermotoga maritima . Bacteria like to flee away from the repellent when high concentrations are present in the environment. CheA is a histidine kinase that associates with CheW, an adaptor protein, will cause the flagella to turn clockwise and result in a tumbling motion. On the other hand, when a high concentration of attractants are present in the environment, the CheA kinase will be turned off, cause flagella to turn counterclockwise, resulting in a forward swimming pattern. [1]
Structure and Functions
Chemoreceptors usually contain a periplasmic ligand binding domain, the transmembrane domain, a HAMP domain which is for signal conversion, and the cytoplasmic domain that contains the methylation sites, flexible bundle and protein binding sites where CheA and CheW bind. CheA is a histidine kinase that contains five units. The P1 domain is the site of substrate autophosphorylation that associates with kinase P4 domain, P2 is where phosphate transfers to CheY, another response regulator protein, from P1. P3, P4 and P5 are the dimerization, kinase and the receptor-coupling domains. The P3 domain was predicted to interact with CheW which stabilize the interface between P3 and P5. The NMR structure has shown that P5 is proximal to the CheW β barrel (residues 635-660). [2] On the right, is the CheW with CheA P4, P5 domains (orange) with CheA P3, P4, P5 (blue).
Molecular Playground banner: P4, P5 of CheA binding with CheW superimpose with P3, P4 P5 of CheA
|
References
- ↑ Hazelbauer, Falke and Parkinson. "Bacterial chemoreceptors: high-performance signaling in networked arrays." Biochemical Sciences, 2007, 33 (1), 9-19. PMID:[1]
- ↑ Park, Borbat, Gonzalez-Bonet, Bhatnagar, et al. "Reconstruction of the chemotaxis receptor-kinase assembly." Nature Structural and Molecular Biology, April 23, 2006, 13 (5), 400-407. PMID:[2]
Acknowledgement
To Luis E Ramirez-Tapia his advice to develop this page.