Sandbox christian
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
The <scene name='Sandbox_christian/Active_site/1'>Active Site</scene> consists of '''Ser 105''', '''Asp 187''' and '''His 224'''. Click here to see the <scene name='Sandbox_christian/Active_site/2'> B-factors</scene> or the <scene name='Sandbox_christian/Active_site/3'>Glycosylation site</scene>. | The <scene name='Sandbox_christian/Active_site/1'>Active Site</scene> consists of '''Ser 105''', '''Asp 187''' and '''His 224'''. Click here to see the <scene name='Sandbox_christian/Active_site/2'> B-factors</scene> or the <scene name='Sandbox_christian/Active_site/3'>Glycosylation site</scene>. | ||
| - | + | ||
</StructureSection> | </StructureSection> | ||
| + | |||
| + | [[Image:1-s2 0-S1381117712001348-gr2.jpg]] | ||
| + | [[Image:1-s2 0-S1381117712001348-fx1.jpg]] | ||
<references/> | <references/> | ||
Revision as of 09:23, 6 September 2012
Candida antarctica lipase B
| |||||||||||
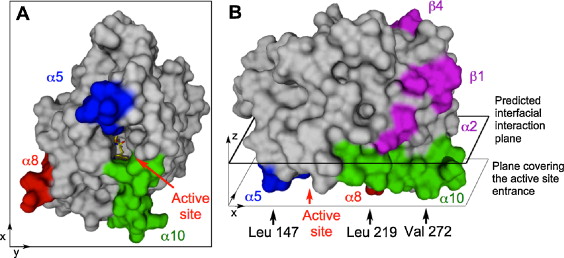 Image:1-s2 0-S1381117712001348-fx1.jpg
Image:1-s2 0-S1381117712001348-fx1.jpg
- ↑ Gruber CC, Pleiss J. Molecular modeling of lipase binding to a substrate-water interface. Methods Mol Biol. 2012;861:313-27. PMID:22426727 doi:10.1007/978-1-61779-600-5_19
- ↑ Gruber CC, Pleiss J. Systematic benchmarking of large molecular dynamics simulations employing GROMACS on massive multiprocessing facilities. J Comput Chem. 2011 Mar;32(4):600-6. doi: 10.1002/jcc.21645. Epub 2010 Sep 1. PMID:20812321 doi:10.1002/jcc.21645
- ↑ Unknown PubmedID 10.1016
