User:Josie N. Harmon/Sandbox 1
From Proteopedia
< User:Josie N. Harmon(Difference between revisions)
| Line 3: | Line 3: | ||
== '''Xanthine Oxidoreductase''' == | == '''Xanthine Oxidoreductase''' == | ||
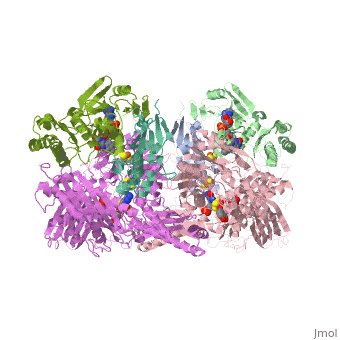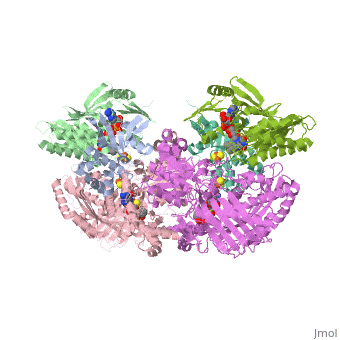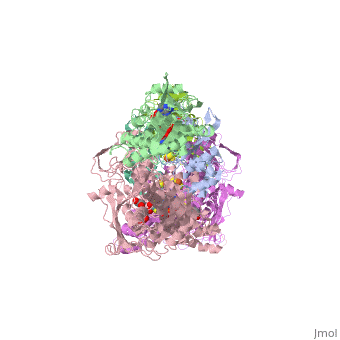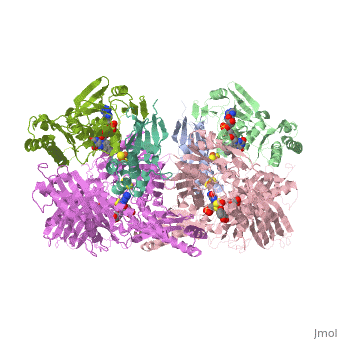
| - | Xanthine oxidase is considered a component of <scene name='User:Josie_N._Harmon/Sandbox_1/Xanthine_oxidoreductase/ | + | Xanthine oxidase is considered a component of <scene name='User:Josie_N._Harmon/Sandbox_1/Xanthine_oxidoreductase/2'>xanthine oxidoreductase</scene> along with xanthine dehydrogenase, which is an enzyme known to generate reactive oxygen species. This enzyme is considered to be extremely important in the catabolism of purines in several steps to yield <scene name='User:Josie_N._Harmon/Sandbox_1/Uric_acid/1'>uric acid</scene> which is ultimately excreted from the body, through a series of oxidation steps than involve metabolizing hypoxanthine to xanthine, which is further oxidized to uric acid. The xanthine oxidoreductase enzyme can exists in two different states, one being the xanthine oxidase conformer and the other being the xanthine dehydrogenase conformer. There are several disulfide bridges within the oxidoreductase enzyme and if these bridges are left intact the enzyme acts as an oxidase, but if these bridges are cleaved the enzyme acts as a dehydrogenase. Also the oxidoreductase enzyme can be permanently cleaved by proteases so that it always acts in the oxidase form. One side of the xanthine oxidoreductase enzyme consists of an <scene name='User:Josie_N._Harmon/Sandbox_1/Catalytic_site/2'>active site </scene>that includes a molybdenum atom which binds to a purine substrate and adds a hydroxyl group. During this process electrons are extracted and funneled from the active site through a string of iron-sulfur clusters to the opposing side of the enzyme. The opposing side then transfers the electrons to NAD or oxygen depending on the dehydrogenase or oxidase nature of the enzyme. One of the final steps in the electron transfer funnels electrons to a FAD group. The dehydrogenase form of the enzyme transfers these electrons to NAD, while the oxidase form blocks NAD through a loop of protein that covers the FAD molecule allowing smaller oxygen molecules to accept the electrons. A <scene name='User:Josie_N._Harmon/Sandbox_1/6mercaptopurine_ligand/3'>secondary depiction</scene> of the structure is represented with purple arrows representing the alpha helix structures and gold arrows representing the beta strand structures. The <scene name='User:Josie_N._Harmon/Sandbox_1/Mercap_helix_blue/1'>helices </scene>of the protein backbone can also be noted by the blue arrows on the structure. The xanthine oxidase complex consists of numerous interactions such as <scene name='User:Josie_N._Harmon/Sandbox_1/Fad_h_bond/2'>hydrogen bonding</scene>, pi-pi interactions, and hydrophobic interactions. |
== '''6-Mercaptopurine''' == | == '''6-Mercaptopurine''' == | ||
Current revision
Xanthine Oxidase
| |||||||||||