We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Fragment-Based Drug Discovery
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
---- | ---- | ||
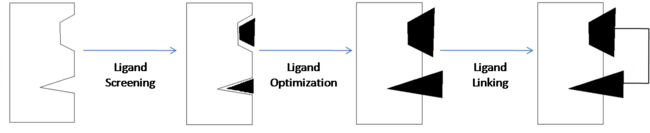
| - | '''Fragment-based drug discovery''' (FBDD) is a method of discovering new compounds by utilizing fragments that have some degree of binding affinity for a drug target, optimizing those fragments so as to increase their binding affinity, then linking them together to form a lead compound that has high affinity and selectivity for the drug target. Nuclear magnetic resonance (NMR) and x-ray crystallography can be used to analyze the fragments and drug targets in order to create three-dimensional images which can be used to obtain an analysis of molecular relationships. This allows developers to get a visual representation of how each fragment binds to the target | + | '''Fragment-based drug discovery''' (FBDD) is a method of discovering new compounds by utilizing fragments that have some degree of binding affinity for a drug target, optimizing those fragments so as to increase their binding affinity, then linking them together to form a lead compound that has high affinity and selectivity for the drug target. Nuclear magnetic resonance (NMR) and x-ray crystallography can be used to analyze the fragments and drug targets in order to create three-dimensional images which can be used to obtain an analysis of molecular relationships. This allows developers to get a visual representation of how each fragment binds to the target and can also be useful in identifying the individual binding sites of the target. |
[[Image:SAR by NMR Illustrated.png | thumb | center | 650px | Fragment-Based Drug Discovery (Adapted from Fig. 1)<ref name="Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.">Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.</ref>]] | [[Image:SAR by NMR Illustrated.png | thumb | center | 650px | Fragment-Based Drug Discovery (Adapted from Fig. 1)<ref name="Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.">Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.</ref>]] | ||
Revision as of 18:58, 31 October 2012
Drug Design: Fragment-Based Drug Discovery
| |||||||||||
References
- ↑ 1.0 1.1 Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.
- ↑ Oltersdorf T., Elmore S. W., Shoemaker A. R. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Vol 435|2 June 2005|doi:10.1038/nature03579
- ↑ Pandit D. LIGAND-BASED DRUG DESIGN: I. CONFORMATIONAL STUDIES OF GBR 12909 ANALOGS AS COCAINE ANTAGONISTS; II. 3D-QSAR STUDIES OF SALVINORIN A ANALOGS AS εΑΡΡΑ OPIOID AGONISTS. http://archives.njit.edu/vol01/etd/2000s/2007/njit-etd2007-051/njit-etd2007-051.pdf