This tutorial is designed for High School (ages 14-19)
[1]
Purpose of the Tutorial
- This tutorial is intended as a beneficial learning/teaching aid for an entry-level chemistry student. Applying general chemistry to a research article will allow the students to see the impact they can have on the research world in the future by applying their knowledge. This tutorial is based on learning, comprehending and applying their knowledge. Various general chemistry topics are discussed in detail for an entry-level student and then shown through various interactive representations of the compound used by the research article.
Summary: Scientific Research Article
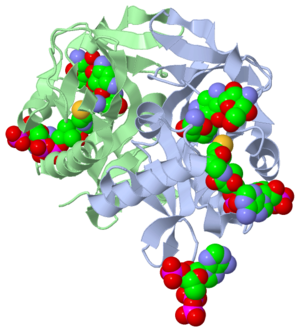
This molecule to right is from the article "Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin" published in Nature Structural Biology. [2]. The study focused on aminoglycoside 2’- N- acetyltransferase (AAC (2’)- Ic) an enzyme. An enzyme is a compound that speeds the rate of a reaction.
The scientists involved in the study determined the structural orientation of AAC (2’)-Ic from Mycobacterium tuberculosis, a pathogen. The specific structure, protein fold, of AAC (2’)-Ic, is placed in the GCN5-related N-acetyltransferase superfamily (GNAT). The protein fold is important because it determines the function of a compound. The GNAT superfamily is a group of enzymes that are similar in structure. Due to the similarity in structure, they also have similar functions.[2]
Although the physiological function of AAC(2’)-Ic is not certain, the structure determined by the scientists allowed them to hypothesize the function. The AAC(2’)-Ic enzyme is located within the mycothiol (a component of the pathogen) structure. AAC(2’)-Ic may be capable of acetylating the aminoglycoside antibiotic. An acetylation is the addition of CH3CO onto a compound, which in this case is the antibiotic. When this occurs the aminoglycoside antibiotic becomes inactive. The basis of this study is important because when pathogens become resistant or inactive to commonly used antibiotics, an infection that used to be easily cured can now become severe and life threatening.[2]
Objectives
By the end of this tutorial you should be able to:
1. Describe and provide examples of covalent bonds, ionic bonds, and hydrogen bonds
2. Understand and identify the importance of secondary structures
3. Understand and describe an active site
4. Describe what a ligand is
5. Understand and explain the importance of Tobramycin as an antibiotic
6. Describe the general function of CoA
7. Be able to classify amino acids and understand what the classification represents
Types of Bonds
There are three common types of bonds. These bonds include hydrogen bonds, covalent bonds, and ionic bonds. The strongest bond is a covalent bond, followed by the ionic bond, which leaves the weakest bond to be the hydrogen bond. Knowing where these bonds are utilized will aid in your understanding of why a structure is in a certain conformation.
Covalent Bonds
The strongest type of bond is the covalent bond. Covalent bonds involve the sharing of electrons between two molecules/atoms. An example of a covalent bond is shown here. This is the amino acid Arginine. Amino Acids are discussed in a later section; this representation is only used to show a covalent bond. The grey molecules are carbons, the blue molecules are nitrogen’s and the red atom is oxygen. All of these atoms are covalently bound. The carbons, nitrogen’s, and oxygen are all sharing electrons with their adjacent atoms. These bonds are very stable and are not easy to break.
Ionic Bonds
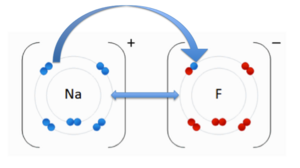
An ionic bond is an attraction between two molecules of opposite charge. The opposite charges are positive (+) and negative (-). A positively charged atom is referred to as a cation, and a negatively charged atom is referred to as an anion. In the image to the right, you see an anion, Fluorine (F) and the cation, Sodium (Na). These two atoms are attracted to each other due to their opposite charges. The double-sided arrow between them is representation of their attractive force. This representation highlights an ionic interaction between Tobramycin and Aspartic acid (Asp). The nitrogen on Tobramycin has a (+) positive charge and Aspartic acid has a (-) negative charge. The opposing charges are attracted to each other forming a ionic bond.
Hydrogen Bonds
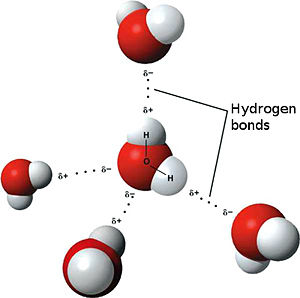
The weakest bond, the hydrogen bond is an attractive interaction between an electronegative atom and hydrogen. Electronegative atoms have high electron density. They are strong atoms that pull electrons towards them from weaker/low electron density atoms, such as hydrogen. When the electronegative atom pulls the electrons, it leaves the other atom with a slightly positive charge. A common example of this is water. The image to the left shows this interaction. The highly electronegative oxygen pulls the hydrogen closer by attracting hydrogen’s electrons. When oxygen pulls the electrons, it leaves hydrogen with a slight positive charge. Since oxygen is pulling the hydrogen’s inward, the formation of a water droplet is possible. In this representation the hydrogen bonds are represented as yellow-dashed lines. The hydrogen bonds are important in this study and this molecular compound because they offer the stability of the secondary structures.
Secondary Structures
Secondary structures are alpha helices and beta sheets. The helices and sheet provide stability to the molecule as a whole. The alpha helices are represented with pink arrows and the beta strands are represented with yellow arrows. This molecule has approximately eight alpha helices and four beta sheets. Alpha helices have a cylinder-like structure that rotates in a clockwise manner with a parallel formation. This representation shows these key features of alpha helices. Here, you can see the helices rotating clockwise (follow the arrows), and the parallel formation within the cylinder structure. The parallel alpha helices are held in its cylinder structure by hydrogen bonds. Beta sheets are often anti-parallel, which are clearly represented in this figure. The folding of a protein, alpha helices and beta sheets, is what gives the compound its function. When there is a change in protein folding, the function will change. From the article summary you know that AAC(2’) has a similar fold to that of the GNAT superfamily. The GNAT fold described in the study has the function of acetylation, the addition of an acetyl group. An acetyl functional group is composed of CH3CO. It is important to note that the discovery of the GNAT fold lead to the understanding of the function of AAC(2’), because of their similar structure. The reaction centered below is the acetylation that occurs to the aminoglycoside antibiotic causing its inactivity. The Acetylation was reconstructed and modified from “Aminoglycoside 2’ –N- Acetyltransferase from Mycobacterium tuberculosis in complex with Coenzyme A and aminoglycoside substrate”, the research article we have been referencing. From this reaction you see the aminoglycoside antibiotic (Ribostamycin) being acted upon by the enzyme AAC(2’). AAC(2’) is adding and acetyl group to the antibiotic. On the right side of the arrow you can see the final product of the acetylation, the antibiotic and acyl group bound. The Acetyl group is circled, so you are able to locate it throughout the reaction. Acetylation is one of the more common reactions that occurs pathologically and physiologically.[2]
Active Site
The active site of a molecule can be described as a pocket where an interaction between compounds occurs. This interaction will cause a change in structure shape. The conformational change, change in structure shape, can inhibit or activate the physiological/pathological affect. The active site is where the ligand is going to bind. (Ligands are discussed in detail in the next section labeled “Ligands”) The active site can either be inhibited or activated by ligands. Referring back to our article, the active site is where the acetylation is going to occur. In this depiction of the active site you can see the pocked where CoA will aid AAC (2’) in the acetylation of Tobramycin (Toy), the aminoglycoside antibiotic. This acetylation at the active site will cause the antibiotic to be inactive, hence inhibiting the active site. When Tobramycin becomes inactivated it is no longer able to aid in the destruction of bacteria. This is what we call antibiotic resistance.
Ligand
Ligands are molecules or complexes that are within the secondary structures that orient in such a way to contribute the function of the complex as a whole. Ligands can have binding sites on receptors, and when bound can trigger a physiological response. A ligand can be a competitive agonist, allosteric agonist, competitive antagonist, or an allosteric antagonist. An agonist is a ligand that causes a physiological response, activating the active site. An antagonist is a ligand that inhibits a physiological response, not allowing the active site to be activated.
A ligand is competitive when it is binding to the same site as the physiological activator; hence it is competing for the same site. When a ligand binds to an allosteric site, the ligand is binding to the same receptor but it is not binding to the active site. The ligands present in the complex used by the research article are coenzyme A, Tobramycin and Phosphate-Adenosine-5'-Diphosphate.
Coenzyme A
Coenzyme (CoA) is a coenzyme that synthesizes and oxidizes fatty acids. This process is essential for the utilization of fatty acids. Coenzyme A is used as a substrate in the citric acid cycle. The citric acid cycle is also known as the Krebs cycle or tricarboxylic acid cycle (TCA). This process is important to the production of ATP, which is an energy source used by the body.
The Protein’s in this molecule are represented as a dimer. A dimer is a chemical structure formed from two identical subunits. Some molecules are present as a dimer because it is more stable then the monomer. The dimer is constructed by connecting two subunits along their axis.
Tobramycin
Tobramycin is an antibiotic part of the aminoglycoside family. Aminoglycosides produce antibacterial effects by inhibiting protein synthesis and compromising the cell wall structure. By inhibiting the protein synthesis of the bacteria, it does not allow the bacteria to replicate. The cell wall is an important structure to bacteria because it provides the structure and stability to the bacteria. By disrupting the cell wall, we are removing the stability of the bacteria and ultimately casing bacteria death.
Tobramycin targets a variety of bacteria, particularly gram(-) species. Just like all drugs there are side effects associated with tobramycin. Some of the more common side effects are ototoxicity and nephrotoxicity. Ototoxic is hearing loss and nephrotoxic is causing kidney damage. The kidney damage is due to Tobramycin reabsorption through the renal tubules. This basically means that tobramycin may be toxic to the kidneys. The toxicity is caused by the contact-time in the renal tubules where the drug is located.
Tobramycin trade name is Tobrex. A trade name is another name for tobramycin. It is a pregnancy category D. Pregnancy categories are assigned to all drugs. They are used to classify how likely the drug is to cause harm to the fetus. The pregnancy categories are A, B, C, D, and X. Pregnancy category A causes no harm to the fetus and pregnancy category X indefinitely causes harm to the fetus. Since Tobramycin is a pregnancy category D, this is not an optimal choice for a pregnant patient.
Tobramycin can be given intravenously, intramuscularly, as an inhalation or ophthalmicly. Intravenously is an IV route of administration where the drug is administered directly to the vasculature or blood vessels. Intramuscular is a shot that penetrates your muscle. A common example of an intramuscular administration would be a flu shot. Inhalation is a route of administration where the lungs are the targets. An example of this would be an inhaler used in asthmatics. Ophthalmic administration is where the drug is administered to the eye; an example would be an eye drop.
Amino Acids
Amino acids are the building blocks of proteins. There are 20 common amino acids. The basic structure of an amino acid is an amine group (-NH2), a carboxylic acid group (-COOH) and a functional group specific to each amino acid. The functional group determines how the amino acid is categorized. They are categorized as polar, non-polar, acidic or basic. Amino acids are commonly referred to using their three-letter abbreviation. I have provided the three-letter abbreviations along side the amino acids in their category list.
An amino acid is considered polar when the charges electronegativity between atoms in the functional group are extremely different. For example, water is considered polar. This is because the electronegativity of oxygen is extremely high and the electronegativity of hydrogen is extremely low. The opposing electronegativity is making water polar. In contrast, non-polar is when the electronegativity of the atoms in the functional group are similar. An amino acid is considered polar when it has an acidic functional group, such as a carboxylic acid (pH~1-6). In correlation, a basic amino acid has a basic functional group, such as an amine group (pH~8-14). An amino acid is neutral when it has a functional group that displays a pH close to physiological pH (~7). A list of the most common amino acids and their categories are as follows for your reference.
Neutral Amino Acids:
Alanine (ala)
Asparagine (asn)
Cysteine (cyc)
Glutamine (gln)
Glycine (gly)
Isoleucine (ile)
Leucine (leu)
Methionine (met)
Phenylalanine (phe)
Proline (pro)
Serine (ser)
Threonine (thr)
Tryptophan (trp)
Tyrosine (tyr)
Valine (val)
Acidic amino acids:
Aspartic acid (asp) Glutamic acid (glu)
Basic Amino acids:
Arginine (arg)Histidine (his) Lysine (lys)
Polar Amino acids:
Arginine (arg) Asparagine (asn) Aspartic acid (asp)Cysteine (cyc) Glutamic Acid (glu) Glutamine (gln) Histidine (his) Lysine (lys) Serine (ser) Threonine (thr) Tryptophan (trp) Tyrosine (tyr)
Non-polar amino acids:
Alanine (ala) Glycine (gly) Isoleucine (ile) Leucine (leu) Methionine (met) Phenylalanine (phe) Proline (pro) Valine (val)
There are 8 different amino acids present in the research article main compound.
CoA Amino Acids
Coenzyme A (CoA) has seven amino acids bound to it. The amino acids consist of two arginine, one glycine, and four valine’s.
Arginine is a basic amino acid and is a nonessential A nonessential amino acid is an amino acid that can be synthesized by the body, so it is not essential that you obtain the amino acid form another source. Glycine is a non-polar, basic amino acid. It is also the smallest common amino acid. The functional group attached is hydrogen. Valine is also categorized as a nonpolar, basic amino acid. Valine is nonpolar and basic because the functional group consists of two methyl groups.
CoA and Arg124
CoA and Gly92
CoA and Val96
CoA and Val86
CoA and Val94
CoA and Val84
Tobramycin Amino Acids
Tobramycin has four amino acids bound to it. There are two aspartic acid, a serine, and a tryptophan.
Asp35
Trp181
Ser117
Asp152
PAP Amino Acids
PAP has four amino acids bound to it. There are two histidine’s and a tryptophan.
His54
Trp90
His55
http://www.elmhurst.edu/~chm/vchembook/561aminostructure.html