Tutorial:Basic Chemistry Topics
From Proteopedia
| Line 82: | Line 82: | ||
='''Secondary Structures'''= | ='''Secondary Structures'''= | ||
| - | Secondary structures are alpha helices and beta sheets. The helices and sheet provide stability to the molecule as a whole. The alpha helices are represented with pink arrows and the beta strands are represented with yellow arrows. This molecule has approximately eight alpha helices and four beta sheets. Alpha helices have a cylinder-like structure that rotates in a clockwise manner with a parallel formation. This representation shows these key features of alpha helices. Here, you can see the helices rotating clockwise (follow the arrows), and the parallel formation within the cylinder structure. The parallel alpha helices are held in its cylinder structure by hydrogen bonds. Beta sheets are often anti-parallel, which are clearly represented in this figure. The folding of a protein, alpha helices and beta sheets, is what gives the compound its function. When there is a change in protein folding, the function will change. | + | Secondary structures are alpha helices and beta sheets. The helices and sheet provide stability to the molecule as a whole. The alpha helices are represented with pink arrows and the beta strands are represented with yellow arrows. This molecule has approximately eight alpha helices and four beta sheets. Alpha helices have a cylinder-like structure that rotates in a clockwise manner with a parallel formation. This representation shows these key features of alpha helices. Here, you can see the helices rotating clockwise (follow the arrows), and the parallel formation within the cylinder structure. The parallel alpha helices are held in its cylinder structure by hydrogen bonds. Beta sheets are often anti-parallel, which are clearly represented in this figure. The folding of a protein, alpha helices and beta sheets, is what gives the compound its function. When there is a change in protein folding, the function will change. |
<scene name='Tutorial:Basic_Chemistry_Topics/Alpha_beta_2ndstructures/1'>Alpha Helices and Beta Sheets</scene> | <scene name='Tutorial:Basic_Chemistry_Topics/Alpha_beta_2ndstructures/1'>Alpha Helices and Beta Sheets</scene> | ||
| Line 94: | Line 94: | ||
[[Image:Recon of acetylation.png | thumb | center | 600px | Visual Representation of an Acetylation<ref name="Article" />]] | [[Image:Recon of acetylation.png | thumb | center | 600px | Visual Representation of an Acetylation<ref name="Article" />]] | ||
| + | From the article summary you know that AAC(2’) has a similar fold to that of the GNAT superfamily. The GNAT fold described in the study has the function of acetylation, the addition of an acetyl group. An acetyl functional group is composed of CH3CO. It is important to note that the discovery of the GNAT fold lead to the understanding of the function of AAC(2’), because of their similar structure. The reaction centered below is the acetylation that occurs to the aminoglycoside antibiotic causing its inactivity. The Acetylation was reconstructed and modified from “Aminoglycoside 2’ –N- Acetyltransferase from Mycobacterium tuberculosis in complex with Coenzyme A and aminoglycoside substrate”, the research article we have been referencing. From this reaction you see the aminoglycoside antibiotic (Ribostamycin) being acted upon by the enzyme AAC(2’). AAC(2’) is adding and acetyl group to the antibiotic. On the right side of the arrow you can see the final product of the acetylation, the antibiotic and acyl group bound. The Acetyl group is circled, so you are able to locate it throughout the reaction. Acetylation is one of the more common reactions that occurs pathologically and physiologically.<ref name="Article" /> | ||
='''Active Site'''= | ='''Active Site'''= | ||
Revision as of 02:14, 8 November 2012
Purpose of the Tutorial
- This tutorial is intended as a beneficial learning/teaching aid for an entry-level chemistry student. Applying general chemistry to a research article will allow the students to see the impact they can have on the research world in the future by applying their knowledge. This tutorial is based on learning, comprehending and applying their knowledge. Various general chemistry topics are discussed in detail for an entry-level student and then shown through various interactive representations of the compound used by the research article.
Summary: Scientific Research Article
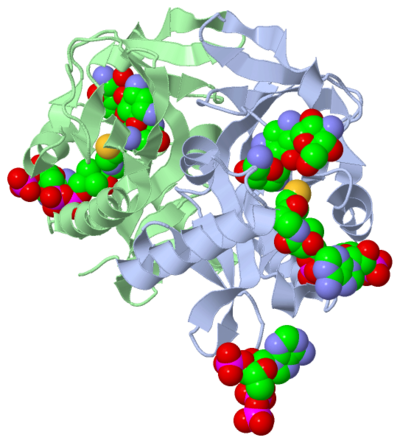
This molecule to right is from the article "Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin" published in Nature Structural Biology. [2]. The study focused on aminoglycoside 2’- N- acetyltransferase (AAC (2’)- Ic), an enzyme. An enzyme is a compound that speeds the rate of a reaction.
The scientists involved in the study determined the structure of AAC (2’)-Ic from Mycobacterium tuberculosis, a pathogen. The specific structure, protein fold, of AAC (2’)-Ic, is placed in the GCN5-related N-acetyltransferase (GNAT) superfamily. The GNAT superfamily is a group of enzymes that are similar in structure. The structure, protein fold, is important because it determines the function of a compound. Since the GNAT superfamily and AAC(2’)-Ic have a similar structures, they also have similar functions.[2]
Although the physiological function of AAC(2’)-Ic is not certain, the structure determined by the scientists allowed them to hypothesize the function. The AAC(2’)-Ic enzyme is located within the mycothiol (a component of the pathogen) structure. AAC(2’)-Ic may be capable of acetylating the aminoglycoside antibiotic. An acetylation is the addition of CH3CO group onto a compound, which in this case is the antibiotic. When this occurs the aminoglycoside antibiotic becomes inactive. The basis of this study is important because when pathogens become resistant or inactive to commonly used antibiotics, an infection that used to be easily cured can now become severe and life threatening.[2]
| |||||||||||
References
- ↑ Vetting, M. W., et al. "Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin." RCSB Protien DataBase. N.p., 28 Aug.2002. Web. 13 July 2011. http://www.rcsb.org/pdb/explore/explore.do?structureId=1M4D
- ↑ 2.0 2.1 2.2 2.3 2.4 Vetting, Matthew W., et al. "Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin."Nature Structural Biology 9.9 (2002): 653-58. Print.
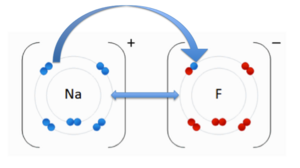
- ↑ . "File:NaF.gif." Wikipedia. Wikipedia, 17 June 2011. Web. 31 Oct. 2012.<http://en.wikipedia.org/wiki/File:NaF.gif.
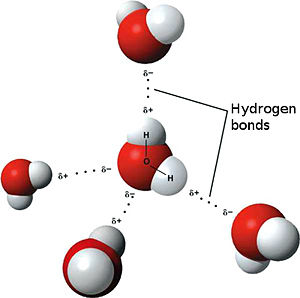
- ↑ Maňas, Michal, trans. "File:3D model hydrogen bonds in water.jpg." Wikimedia Commons. Wikimedia Commons, 3 Dec. 2007. Web. 31 Oct. 2012 <http://commons.wikimedia.org/wiki/File:3D_model_hydrogen_bonds_in_water.jpg.