We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Josie N. Harmon/Sandbox Tutorial
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
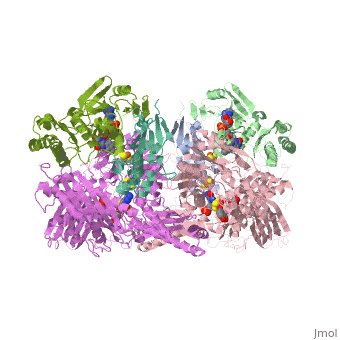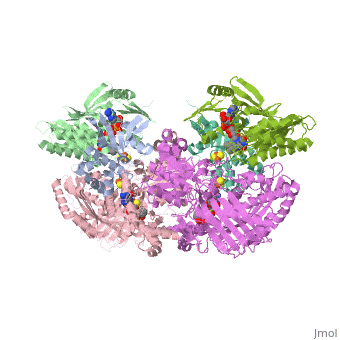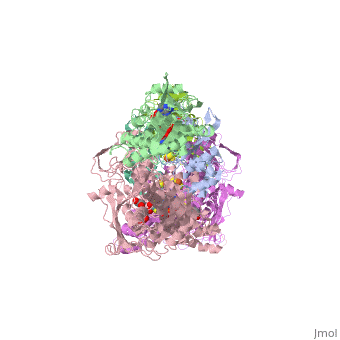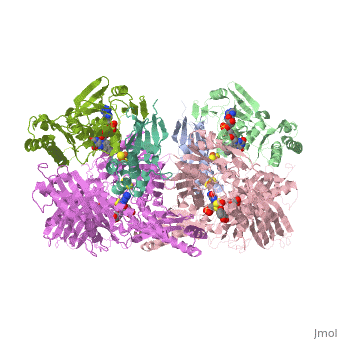
1. Within the oxidoreductase enzyme there are multiple <scene name='Josie_N._Harmon/Sandbox_Tutorial/Disulfide_bonds/3'>disulfide bridges</scene>. In the case that these bridges are left untouched the enzyme will then take on the characteristics to act as an oxidase, but in the event that these <scene name='Josie_N._Harmon/Sandbox_Tutorial/Disulfide_bonds_no/1'>bridges are restricted</scene> the enzyme then acts as a dehydrogenase. | 1. Within the oxidoreductase enzyme there are multiple <scene name='Josie_N._Harmon/Sandbox_Tutorial/Disulfide_bonds/3'>disulfide bridges</scene>. In the case that these bridges are left untouched the enzyme will then take on the characteristics to act as an oxidase, but in the event that these <scene name='Josie_N._Harmon/Sandbox_Tutorial/Disulfide_bonds_no/1'>bridges are restricted</scene> the enzyme then acts as a dehydrogenase. | ||
| - | 2. The oxidoreductase enzyme | + | 2. The oxidoreductase enzyme can be permanently altered by proteases so that it will be constantly utilized in the oxidase form. |
== Mechanism of Action == | == Mechanism of Action == | ||
| Line 22: | Line 22: | ||
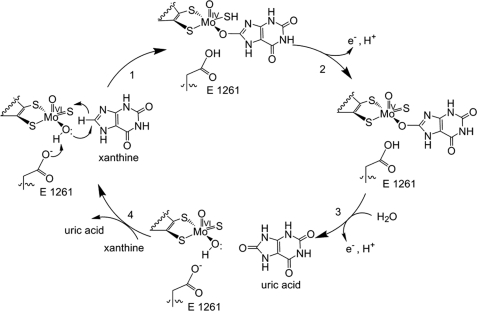
[[Image:XOmech.jpg]] | [[Image:XOmech.jpg]] | ||
| - | Following the intramolecular electron transfer that occurs within the enzyme, the enzyme is able to catalyze the reaction of purine degradation products to ultimately yield uric acid. The comprehensive catalytic activity of the enzyme consists of a redox reaction that includes a reduction reaction in which the substrate is oxidized by a hydroxylation at the molybdenum center and a oxidation reaction in which electrons are removed from the enzyme by it's FAD. During the reduction reaction the substrate is hydrolyzed (at a specific carbon) by a nucleophilic attack with the MO-OH group within the enzyme's metal center. While this nucleophilic attack is occurring, there is also an accompanying step taking place where the carbon being hydroxylized is transferring a hydride to a MO=S group. The bound substrate product is then expelled by solvent hydroxide to regenerate the original MO-OH ligand to be used for consecutive catalytic cycles. The oxidation reaction on the other hand consists of transporting electrons from the FAD center to a terminal electron acceptor such as NAD+ (dehydrogenase) or oxygen (oxidase) | + | Following the intramolecular electron transfer that occurs within the enzyme, the enzyme is able to catalyze the reaction of purine degradation products to ultimately yield uric acid. The comprehensive catalytic activity of the enzyme consists of a redox reaction that includes a reduction reaction in which the substrate is oxidized by a hydroxylation at the molybdenum center and a oxidation reaction in which electrons are removed from the enzyme by it's FAD. During the reduction reaction the substrate is hydrolyzed (at a specific carbon) by a nucleophilic attack with the MO-OH group within the enzyme's metal center. While this nucleophilic attack is occurring, there is also an accompanying step taking place where the carbon being hydroxylized is transferring a hydride to a MO=S group. The bound substrate product is then expelled by solvent hydroxide to regenerate the original MO-OH ligand to be used for consecutive catalytic cycles. The oxidation reaction on the other hand consists of transporting electrons from the FAD center to a terminal electron acceptor such as NAD+ (dehydrogenase) or oxygen (oxidase). |
== Clinical Application == | == Clinical Application == | ||
Revision as of 16:21, 3 December 2012
Xanthine Oxidase Biochemistry Tutorial
The purpose of this tutorial is to explain the mechanism of the metabolic enzyme xanthine oxidoreductase.
| |||||||||||