Catalytic Subunit of T. Castaneum TERT Polymerase
From Proteopedia
(Difference between revisions)
m |
|||
| Line 1: | Line 1: | ||
| - | < | + | <StructureSection load='3kyl' size='500' frame='true' align='right' caption='Catalytic Subunit of TERT Polymerase bound to RNA Promoter and DNA Template Strand' [[3kyl]]> |
| Line 5: | Line 5: | ||
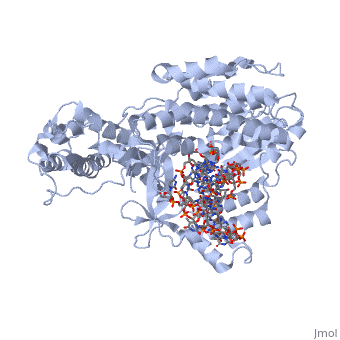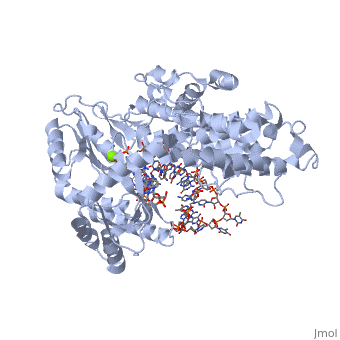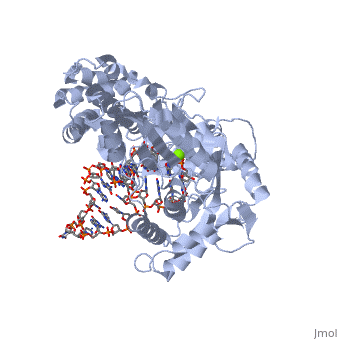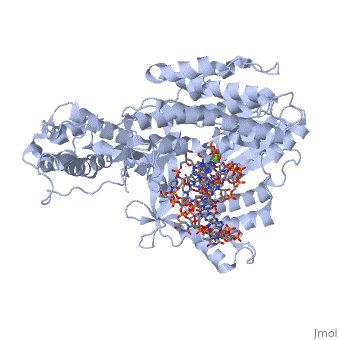
Telomerase is a specialized DNA polymerase that extends the 3' ends of eukaryotic linear chromosomes, a process required for genomic stability and cell viability. Here we present the crystal structure of the active Tribolium castaneum telomerase catalytic subunit, TERT, bound to an RNA-DNA hairpin designed to resemble the putative RNA-templating region and telomeric DNA. The RNA-DNA hybrid adopts a helical structure, docked in the interior cavity of the TERT ring. Contacts between the RNA template and motifs 2 and B' position the solvent-accessible RNA bases close to the enzyme active site for nucleotide binding and selectivity. Nucleic acid binding induces rigid TERT conformational changes to form a tight catalytic complex. Overall, TERT-RNA template and TERT-telomeric DNA associations are remarkably similar to those observed for retroviral reverse transcriptases, suggesting common mechanistic aspects of DNA replication between the two families of enzymes. | Telomerase is a specialized DNA polymerase that extends the 3' ends of eukaryotic linear chromosomes, a process required for genomic stability and cell viability. Here we present the crystal structure of the active Tribolium castaneum telomerase catalytic subunit, TERT, bound to an RNA-DNA hairpin designed to resemble the putative RNA-templating region and telomeric DNA. The RNA-DNA hybrid adopts a helical structure, docked in the interior cavity of the TERT ring. Contacts between the RNA template and motifs 2 and B' position the solvent-accessible RNA bases close to the enzyme active site for nucleotide binding and selectivity. Nucleic acid binding induces rigid TERT conformational changes to form a tight catalytic complex. Overall, TERT-RNA template and TERT-telomeric DNA associations are remarkably similar to those observed for retroviral reverse transcriptases, suggesting common mechanistic aspects of DNA replication between the two families of enzymes. | ||
| + | |||
| + | =Structural Overview= | ||
| + | |||
| + | TERT is a specialized protein belonging to the same family as DNA Polymerase. It shares the same structural motifs with a slightly different arrangement. On the whole polymerases are composed of structurally conserved motifs: the "fingers" "thumb" and "palm". TERT polymerase is similar in structure to retroviral reverse transcriptases such as HIV Reverse transcriptase. However, unlike other transcriptases TERT exists as a stable RNA-Protein complex, with an RNA molecule acting as the template molecule for dNTP addition. It is this RNA primer which allows for the high fidelity and high processivity of TERT Polymerase. | ||
| + | |||
| + | The binding site consists of a highly positively charged pocket that is configured in such a way that the DNA is inserted with its 3' end at the active site for dNTP addition. The structure shown above is a TERT complex with a DNA-RNA hairpin loop hybrid. The hybrid in the crystal structure [http://www.proteopedia.com/wiki/index.php/Image:Hairpin_Loop.png shown here] was developed to determine the exact telomeric repeat sequence of the RNA primer. | ||
| + | |||
| + | ===Major Domains=== | ||
| + | =====Thumb===== | ||
| + | There are four major interaction domains in this protein. The thumb domain of the enzyme is implicated in DNA binding and processivity. The thumb domain, with the active residues Lys416 and Asn423, forms hydrogen bonds with the phosphates in the DNA backbone and keeps the base pairs oriented towards the inside of the binding pocket. A portion of the thumb domain, residues Cys390 and Gly391 are also responsible for guiding the DNA substrate into the binding pocket through bulky interactions and electron repulsion. The thumb domain runs almost completely parallel to the curvature of the DNA substrate. | ||
| + | |||
| + | =====Palm===== | ||
| + | The palm domain contains the catalytic site of the enzyme where the structurally significant Tyr256, Gln308, and Val342 reside. These residues form hydrogen bonds with the 3' end of the DNA and the incoming nucleotides for synthesis of telomeric DNA. | ||
| + | |||
| + | =====Finger===== | ||
| + | |||
| + | =====TRBD===== | ||
| + | |||
| + | |||
| + | ====Positive Binding Pocket==== | ||
| Line 16: | Line 36: | ||
<scene name='Catalytic_Subunit_of_T._Castaneum_TERT_Polymerase/Positive_ionic_binding_pocket/1'>Positive Binding Pocket</scene> | <scene name='Catalytic_Subunit_of_T._Castaneum_TERT_Polymerase/Positive_ionic_binding_pocket/1'>Positive Binding Pocket</scene> | ||
| + | |||
| Line 21: | Line 42: | ||
</StructureSection> | </StructureSection> | ||
| + | |||
| + | [[Image:Hairpin_Loop.png|500px|center]] | ||
Revision as of 21:31, 3 December 2012
| |||||||||||