Catalytic Subunit of T. Castaneum TERT Polymerase
From Proteopedia
| Line 35: | Line 35: | ||
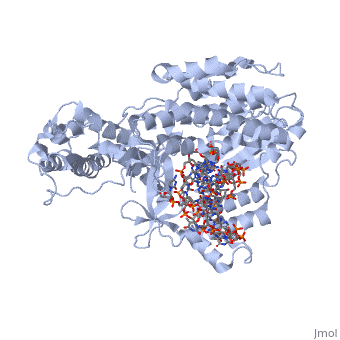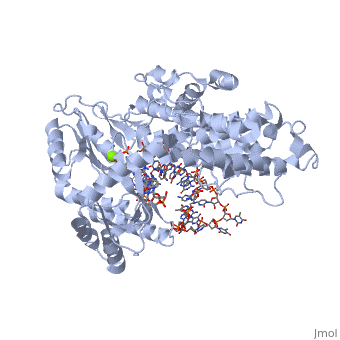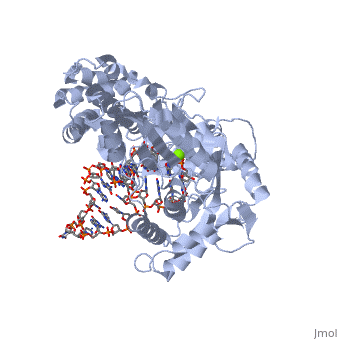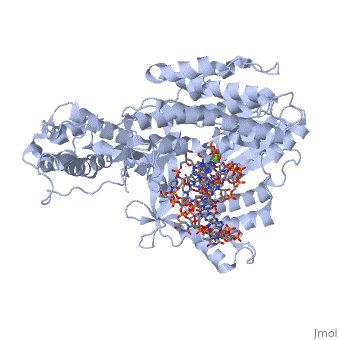
The TERT RNA Binding Domain is the portion of the protein that contains the stable, complexed RNA template strand. It is here that the DNA-RNA hairpin loop in the structure binds.<ref name="Main"/> | The TERT RNA Binding Domain is the portion of the protein that contains the stable, complexed RNA template strand. It is here that the DNA-RNA hairpin loop in the structure binds.<ref name="Main"/> | ||
| + | =Enzyme Activity= | ||
| + | ==Repeat Addition Processivity== | ||
| + | TERT Polymerase has the unique ability to add multiple repeat sequences of DNA to the end of chromosomes, a process which is known as Repeat Addition Processivity. This high amount of processivity can be attributed to the N-terminal association of the TERT subunit with the TEN subunit, the RNA templating region. In this structure unfortunately we do not see the actual association of RNA with the TRBD, however comparisons with other studies and known structural homologies we know that the RNA would bind in such a way as to place the 5' end of the template near the opening of the RNA binding pocket of the TRBD domain. | ||
| + | |||
| + | Because the RNA template has such a high affinity for the TERT molecule, it forces the enzyme to stall once dNTP addition has reached the 5' end of the RNA template. This causes TERT to dissociate from the expanding DNA chain and re-attach for another round of telomeric addition. | ||
| + | |||
| + | ==Conformational Changes== | ||
| + | Comparisons between the substrate bound TERT molecule and the substrate free molecule show small rigid body conformational changes in the positions of the subunits of the enzyme. These small conformational changes translate to a 3.5 angstrom decrease in the diameter of the interior cavity of the closed ring. The precise role of this conformational change is not known, although it may have to do with RNA binding ''in vivo'' | ||
</StructureSection> | </StructureSection> | ||
| Line 43: | Line 51: | ||
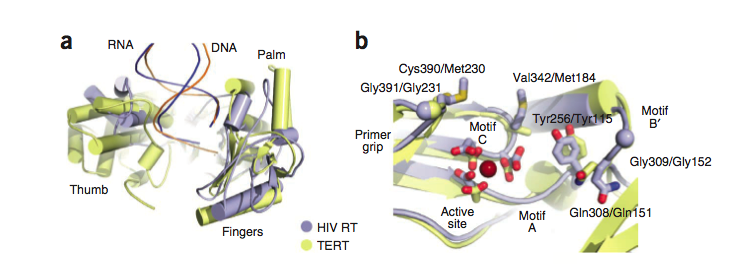
| - | ===Structural Similarity to Reverse Transcriptase=== | + | ===Structural Similarity to HIV Reverse Transcriptase=== |
It has been postulated that the mechanism by which TERT performs elongation of the telomeres is similar to the mechanism used by reverse transcriptases such as those found in retroviruses like HIV. Comparing the two proteins, it can be seen that there exist subtle differences in the structure, but the residues of the active sites are highly conserved across the two types of proteins, and the structural domains remain the same across the two as well. For example, the thumb loop of both is responsible for guiding the DNA substrate into the binding pocket of the enzyme, and orienting it in the correct direction. The Tyr256 and Gln308 of the nucleotide binding pocket are also found in HIV-RT. <ref name="Main"/> | It has been postulated that the mechanism by which TERT performs elongation of the telomeres is similar to the mechanism used by reverse transcriptases such as those found in retroviruses like HIV. Comparing the two proteins, it can be seen that there exist subtle differences in the structure, but the residues of the active sites are highly conserved across the two types of proteins, and the structural domains remain the same across the two as well. For example, the thumb loop of both is responsible for guiding the DNA substrate into the binding pocket of the enzyme, and orienting it in the correct direction. The Tyr256 and Gln308 of the nucleotide binding pocket are also found in HIV-RT. <ref name="Main"/> | ||
Revision as of 21:25, 4 December 2012
Contents |
Introduction
Telomerase is a specialized DNA polymerase that extends the 3' ends of eukaryotic linear chromosomes, a process required for genomic stability and cell viability. Here we present the crystal structure of the active Tribolium castaneum telomerase catalytic subunit, TERT, bound to an RNA-DNA hairpin designed to resemble the putative RNA-templating region and telomeric DNA. The RNA-DNA hybrid adopts a helical structure, docked in the interior cavity of the TERT ring. Contacts between the RNA template and motifs 2 and B' position the solvent-accessible RNA bases close to the enzyme active site for nucleotide binding and selectivity. Nucleic acid binding induces rigid TERT conformational changes to form a tight catalytic complex. Overall, TERT-RNA template and TERT-telomeric DNA associations are remarkably similar to those observed for retroviral reverse transcriptases, suggesting common mechanistic aspects of DNA replication between the two families of enzymes. [1]
| |||||||||||
|
Structural Similarity to HIV Reverse Transcriptase
It has been postulated that the mechanism by which TERT performs elongation of the telomeres is similar to the mechanism used by reverse transcriptases such as those found in retroviruses like HIV. Comparing the two proteins, it can be seen that there exist subtle differences in the structure, but the residues of the active sites are highly conserved across the two types of proteins, and the structural domains remain the same across the two as well. For example, the thumb loop of both is responsible for guiding the DNA substrate into the binding pocket of the enzyme, and orienting it in the correct direction. The Tyr256 and Gln308 of the nucleotide binding pocket are also found in HIV-RT. [1]
Current Research
Aging Research
Because the shortening of the telomere is directly responsible for the eventual senescence of the cell and by extension the aging of the individual, research on effectively elongating the telomere either by introducing synthetic TERT protein or by stimulating the natural production of TERT by the cells is a biological study of great significance.
In 2012 researchers at the Spanish National Cancer Research Centre (CNIO) showed a 24% increase in lifespan for mice who were injected with a virus containing the genes for telomerase production. Telomerase is actually expressed in most in vivo cells, however its levels are not high enough to completely combat the effects of the damage done by replication. With the addition of Telomerase genes by an adeno-associated Virus (AAV) late in the life of the mouse a longer lifespan was observed. Whereas previous studies have shown the possibility of altering telomerase genes during embryonic development to elongate the lifespan, this is the first study to show that an in vivo gene therapy can be effective in doing the same.
The results also showed no higher negligibly higher instances of cancer in these mice, as has been observed in several other studies wherein telomerase is reactivated. In addition to this, a slight rejuvenating effect was seen, with metabolism and mitochondrial fitness increasing a considerable amount. [2]
Cancer Research
Telomerase function has been heavily linked with cancer proliferation and as such it is one of the most common markers for cancer testing. Because of this heavy link to cancer, it is a prime target for many types of cancer research.[3]
TERT Polymerase has been shown to be inhibited by the human protein PinX1. PinX1 works by binding to the site where the RNA template is usually bound. The PinX1 protein is usually found in the nucleolus and nucleoplasm in humans, but in cancer cells TERT is found in the nucleoplasm. It is postulated that PinX1 acts as a tumor suppressor for this reason. [4]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010 Apr;17(4):513-8. Epub 2010 Mar 28. PMID:20357774 doi:10.1038/nsmb.1777
- ↑ Bernardes de Jesus B, Vera E, Schneeberger K, Tejera AM, Ayuso E, Bosch F, Blasco MA. Telomerase gene therapy in adult and old mice delays aging and increases longevity without increasing cancer. EMBO Mol Med. 2012 Aug;4(8):691-704. doi: 10.1002/emmm.201200245. Epub 2012 May, 15. PMID:22585399 doi:10.1002/emmm.201200245
- ↑ European Journal of Cancer, Volume 33, Issue 5, April 1997, Pages 787–791 http://dx.doi.org/10.1016/S0959-8049(97)00062-2
- ↑ FEBS Letters, Volume 579, Issue 4, 7 February 2005, Pages 859–862 http://dx.doi.org/10.1016/j.febslet.2004.11.036