Lactate Dehydrogenase
From Proteopedia
| Line 25: | Line 25: | ||
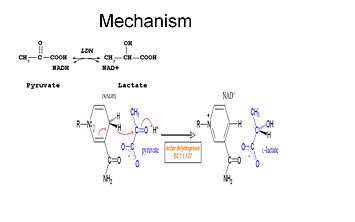
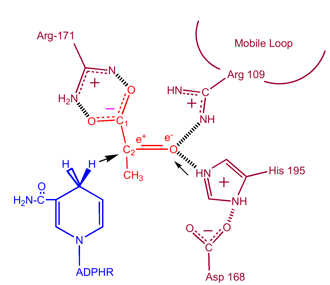
Kinetic studies of lactate dehydrogenase with oxalate and oxamate (structural analogues of lactate and pyruvate)have proven the mechanism stated above. The rate limiting step in this reaction is the rate of dissociation of NAD+ and NADH. The conversion of pyruvate to lactate with the subsequent regeneration of NAD+ is very favorable. | Kinetic studies of lactate dehydrogenase with oxalate and oxamate (structural analogues of lactate and pyruvate)have proven the mechanism stated above. The rate limiting step in this reaction is the rate of dissociation of NAD+ and NADH. The conversion of pyruvate to lactate with the subsequent regeneration of NAD+ is very favorable. | ||
| - | [[Image: | + | [[Image:Lac.PNG]] |
==Regulation== | ==Regulation== | ||
Revision as of 13:24, 10 January 2013
| |||||||||||
Contents |
3D structures of lactate dehydrogenase
Updated June 2012
2e37, 2v6m – TtL-LDH – Thermus thermophilus
2xxe, 3zzn, 4a73– TtL-LDH (mutant)
2x8l, 1ceq – PfL-LDH – Plasmodium falciparum
2zqy, 2zqz, 1llc – LcL-LDH – Lactobacillus casei
3d0o - SaL-LDH – Staphylococcus aureus
2v65 - L-LDH A chain – Champsocephalus gunnari
2v6b – L-LDH – Deinococcus radiodurans
2frm – CpLDH – Cryptosporidium parvum
1sov, 1pze - TgL-LDH – Toxoplasma gondii
1v6a – L-LDH A chain – Cyprinus carpio
1y6j – L-LDH – Clostridium thermocellum
1ldb - GsL-LDH – Geobacillus stearothermophilus
1ldm, 6ldh, 8ldh – sdM4-LDH – spiny dogfish
2ldx – C4-LDH - mouse
3pqe – BsL-LDH (mutant) – Bacillus subtilis
L-LDH from yeast (cytochrome b2)
1kbj - LDH FMN-binding domain
1sze, 1lco, 2oz0 - LDH FMN-binding domain (mutant)
1kbi - LDH FMN-binding domain + pyruvate
1szf, 1ldc - LDH FMN-binding domain (mutant) + pyruvate
1szg - LDH FMN-binding domain + sulfo-FMN
3ks0 - LDH residues 86-180 + flavocytochrome b2 heme domain + FAB B2B4
L-LDH binary complex
2ydn – PfL-LDH + bicine
1cet - PfL-LDH + chloroquine
1t2c - PfL-LDH + NAD
1ez4 - L-LDH + NAD – Lactobacillus pentosus
1lld, 2ldb - GsL-LDH + NAD
2a92 – PvL-LDH + NAD – Plasmodium vivax
2a94, 2aa3 - PvL-LDH + NAD-analog
1u4o, 1u4s, 1u5a, 1xiv – PfL-LDH + inhibitor
2xxb – TtL-LDH (mutant) + AMP
3pqf – BsL-LDH (mutant) + NAD
3vkv - LcLDH + fructose-bisphosphate
4aj1, 4aj2, 4aj4, 4aje, 4ajh, 4aji, 4ajj, 4ajk, 4ajl, 4ajn, 4ajo, 4al4 – LDH α chain + inhibitor – rat
4ajp – hLDH α chain + inhibitor - human
L-LDH ternary complex with inhibitor
1u5c - PfL-LDH + inhibitor + NAD
1t24, 1t25, 1t26 - PfL-LDH + azole derivative + NAD
1t2e - PfL-LDH (mutant) + oxamate + NAD
1ldg - PfL-LDH + oxamate + NAD
2xxj – TtL-LDH (mutant) + oxamate + NAD
2v7p - TtL-LDH + oxamate + NAD
3om9, 3czm – TgL-LDH + OXQ + NAD
3h3f – L-LDH A chain + oxamate + NAD – rabbit
2fnz - CpLDH + oxamate + NAD
1t2f - hL-LDH B chain + azole derivative + NAD
1i0z - hL-LDH H chain + oxamate + NAD
1i10 - hL-LDH M chain + oxamate + NAD
1oc4 – L-LDH + oxamate + NAD – Plasmodium berghei
1a5z - L-LDH + oxamate + NAD – Thermotoga maritima
1lth - L-LDH + oxamate + NAD – Bifidobacterium longum
1ldn - GsL-LDH + oxamate + NAD
9ldb, 9ldt - pL-LDH + oxamate + NAD – pig
L-LDH ternary complex with reactants
1sow, 1pzh - TgL-LDH + oxalate + NAD
1t2d - PfL-LDH + oxalate + NAD
1pzf - TgL-LDH + oxalate + NAD-analog
1pzg - TgL-LDH + sulfate + NAD-analog
3h3j – SaL-LDH (mutant) + pyruvate + NAD
3d4p - SaL-LDH + pyruvate + NAD
2fn7 - CpLDH + lactate + NAD
2ewd - CpLDH + pyruvate + NAD-analog
2fm3 - CpLDH + pyruvate + NAD
5ldh - pLDH + citrate + NAD-analog
3ldh - sdLDH + pyruvate + NAD
3pqd - BsLDH + fructose-bisphosphate + NAD
D-LDH
3kb6 – D-LDH + lactate + NAD – Aquifex aeolicus
1j49 – LdD-LDH + sulfate + NAD – Lactobacillus delbrueckii
1j4a - LdD-LDH + sulfate
1f0x – D-LDH + FAD – Escherichia coli
2dld - D-LDH + oxamate + NAD – Lactobacillus helveticus
L-LDH II
L-Lactate/malate dehydrogenase
1hye, 1hyg – L-LDH/MDH – Methanocaldococcus jannaschi
Additional Information
For additional information, see Carbohydrate Metabolism
Reference
- 1- http://www.bioc.aecom.yu.edu/labs/calllab/highlights/LDH.htm
- 2- http://www.cheric.org/ippage/e/ipdata/2004/05/file/e200405-701.pdf
- 3- http://resources.metapress.com/pdf-preview.axd?code=ulnhp23038060m21&size=largest
- 4- http://www.u676.org/Documents/Chretien-ClinChimActa-95.pdf
- 5- http://www.jbc.org/content/243/17/4526.full.pdf+html
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Ann Taylor, Jasper Small, David Canner