Jms/sandbox8
From Proteopedia
(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | == | + | == Prediction about Hsp12 with Gfp == |
| - | <StructureSection load=' | + | <StructureSection load='2ljl' size='350' side='right' caption='Structure of Hsp12 (PDB entry [[1ljl]])' scene=''> |
| - | + | The GFP likely does not interfere with the native function of Hsp12 because it is far from the localization tags while in the dynamic state in the cytoplasm. And in the membrane it is far from the helixes that actually stabalize the membrane. | |
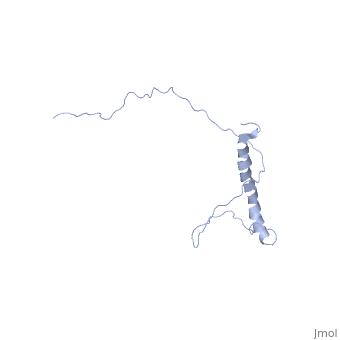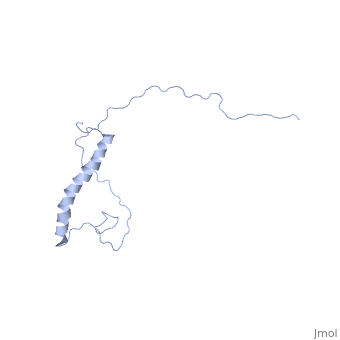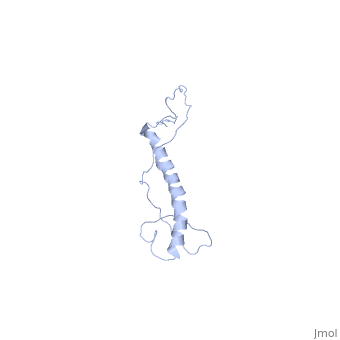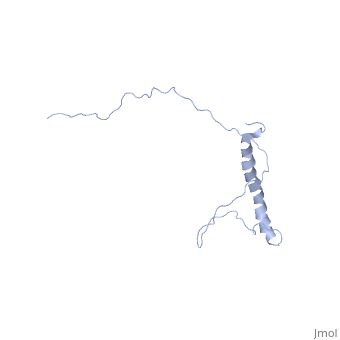
| - | + | In independent studies Hsp12 was shown to form four helixes upon <scene name='Jms/sandbox8/Hsp12_membrane/1'>localization into the membrane of SDS micelles</scene> | |
| + | which was repeated <scene name='Jms/sandbox8/Hsp12_membrane_2/1'>with the same results</scene> | ||
| + | however, in a dodecaphosphatidylcholine (DPC) micelle, it <scene name='Jms/sandbox8/Hsp12_unfolded/1'>only forms one helix</scene> | ||
</StructureSection> | </StructureSection> | ||
| - | <scene name='Jms/sandbox8/Hsp12_unfolded/1'>TextToBeDisplayed</scene> | ||
Current revision
Prediction about Hsp12 with Gfp
| |||||||||||