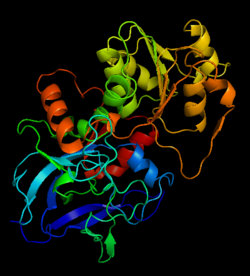
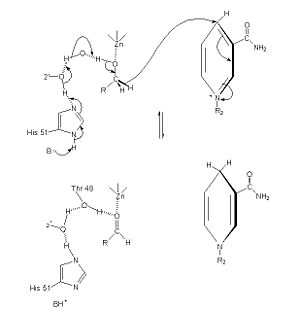
Alcohol dehydrogenase
From Proteopedia
| Line 1: | Line 1: | ||
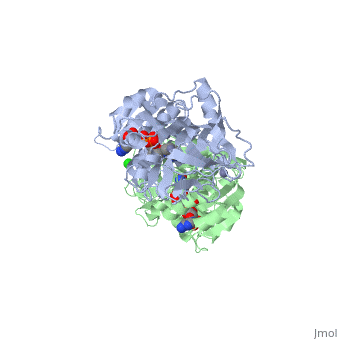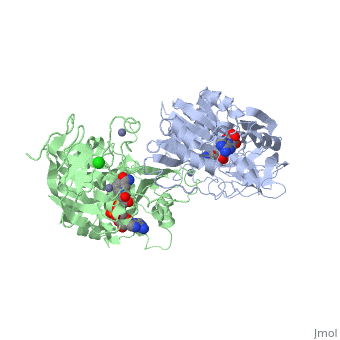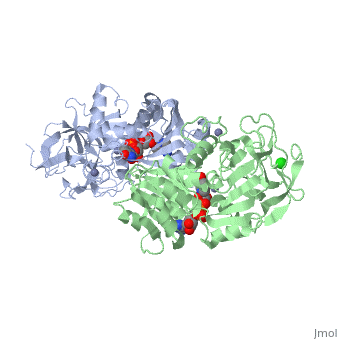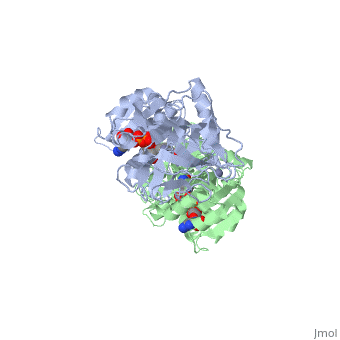
| - | <StructureSection load='1hdz' size='450' side='right' scene='' caption=''> | + | <StructureSection load='1hdz' size='450' side='right' scene='' caption='Human alcohol dehydrogenase dimer with NAD, Ca+2 and Cl- ions (PDB code [[1hdz]])'> |
[[Image:1htb2.png|thumb|left|250px|Structure of Alcohol Dehydrogenase]] | [[Image:1htb2.png|thumb|left|250px|Structure of Alcohol Dehydrogenase]] | ||
Revision as of 09:03, 14 August 2013
| |||||||||||
Additional Resources
For additional information, see: Carbohydrate Metabolism
3D Structures of Alcohol dehydrogenase
Updated on 14-August-2013
ADH I
3jv7 – RrADH I – Rhodococcus rubber
2vna - hADH I catalytic domain - human
2hcy – yADH I – yeast
4eex – LlADH I – Lactococcus lactis
4eez – LlADH I (mutant)
ADH I binary complex
1u3t – hADH I α chain + inhibitor
1hsz, 1hdz, 3hud - hADH I β chain + NAD
1u3w - hADH I γ chain + inhibitor
1ht0 - hADH I γ chain (mutant) + NAD
ADH I ternary complex
2xaa – RrADH I + NAD + alcohol
3fx4 – pADH I + NADP + inhibitor – pig
2w98, 2w4q – hADH I catalytic domain + NADP + inhibitor
1hso - hADH I α chain + NAD + pyrazole derivative
1hdx - hADH I β chain + NAD + alcohol
1u3u, 1u3v - hADH I β chain + inhibitor
1deh, 1hdy - hADH I β chain + NAD + pyrazole derivative
1htb - hADH I β3 chain + NAD + pyrazole derivative
ADH II
3owo – ZmADH II iron-dependent – Zymomonas mobilis
ADH II binary complex
3ox4 - ZmADH II iron-dependent + NAD
3cos - hADH II + NAD + Zn
1e3e – mADH II + NADH – mouse
1e3l - mADH II (mutant) + NADH
1e3i - mADH II + NADH + inhibitor
ADH III
1m6h, 1m6w, 1teh - hADH III χ chain
2fze - hADH III χ chain + ADP-ribose
2fzw - hADH III χ chain + NAD
1mc5 – hADH III χ chain + glutathione + NADH
1ma0 - hADH III χ chain + dodecanoic acid + NAD
3qj5 - hADH III χ chain + inhibitor + NAD
4dl9, 4dlb – tADH III + NAD – tomato
4dla – tADH III
ADH IV
1ye3, 8adh, 5adh - hoADH IV e chain – horse
1qlj - hoADH IV e chain (mutant)
3iv7 – ADH IV – Corynebacterium glutamicum
ADH IV binary complex
2jhf, 2jhg, 1het, 1heu, 1hf3, 1ee2, 2oxi, 2ohx, 6adh - hoADH IV e chain + NAD
1adb, 1adc, 1adf, 1adg, 7adh - hoADH IV e chain + NAD derivative
1mgo, 1ju9, 1qlh, 1a72 - hoADH IV e chain (mutant) + NAD
1d1s, 1agn – hADH IV σ chain + NAD
1d1t - hADH IV σ chain (mutant) + NAD
ADH IV ternary complex
3oq6, 1qv6, 1qv7, 1a71, 1axe, 1axg – hoADH IV e chain (mutant) + NAD + alcohol
4dwv, 4dxh - hoADH IV e chain + NAD + alcohol
1p1r, 1ldy, 1lde - hoADH IV e chain + NADH + formamide derivative
1n92 - hoADH IV e chain + NAD + pyrazole derivative
1bto, 3bto - hoADH IV e chain + NADH + butylthiolane derivative
1n8k - hoADH IV e chain (mutant) + NAD + pyrazole
1mg0, 1hld - hoADH IV e chain + NAD + alcohol
ADH
1a4u – SlADH – Scaptodrosophila lebanonensis
3my7 – ADH ACDH domain – Vibrio parahaemolyticus
3meq – ADH – Brucella suis
3l4p – ADH – Desulfovibrio gigas
1jvb - SsADH – Sulfolobus solfataricus
3i4c, 1nto, 1nvg – SsADH (mutant)
3goh – ADH – Shewanella oneidensis
3gaz – ADH residues 2-334 – Novosphingobium aromaticivorans
2eih – ADH – Thermus thermophilus
1rjw – GsADH – Geobacillus stearothermophilus
1vj0, 1vhd – TmADH -Thermotoga maritima
2eer – ADH – Sulfolobus tokodaii
3uog – ADH – Sinorhizobium meliloti
ADH binary complex
3l77, 3tn7 – ADH short-chain + NADP – Thermococcus sibiricus
1h2b – ADH + NAD – Aeropyrum pernix
1f8f – Benzyl-ADH + NAD – Acinetobacter calcoaceticus
1o2d - TmADH + NADP
1b16, 1b14, 1b15 - SlADH + NAD derivative
1cdo – ADH + NAD - cod
1rhc – ADH F420-dependent +F420-acetone – Methanoculleus thermophilus
1agn – hADH (sigma) +NAD
3pii – GsADH + butyramide
3rj5, 3rj9 – SlADH (mutant) + NAD
3s1l – ADH + Zn – Ralstonia eutropha
ADH ternary complex
1mg5 – ADH + NADH + acetate – Drosophila melanogaster
1r37 – SsADH + NAD + alcohol
1sby – SlADH + NAD + alcohol
1b2l - SlADH + NAD + cyclohexanone
1llu - ADH + NAD + alcohol – Pseudomonas aeruginosa
3cv7 – pADH + NAD + NAP
3rf7 – SoADH + NAD + Fe + Ni
NADP-dependent ADH
1ped - CbADH – Clostridium beijerinckii
2b83, 1jqb – CbADH (mutant)
2nvb - TbADH (mutant) – Thermoanaerobacter brockii
3ftn, 3fpc, 3fpl, 3fsr – ADH chimera
1y9a - EhADH – Entamoeba histolytica
2oui – EhADH (mutant)
1p0c – RpADH8 – Rana perezi
4gac - mADH
NADP-dependent ADH binary complex
1kev – CbADH + NADPH
1bxz – CbADH catalytic domain + alcohol
1ykf – TbADH + NADP
3h4g – pADH + NADP
1p0f – RpADH + NADP
R-specific ADH
1nxq - LbRADH – Lactobacillus brevis
1zk2, 1zk3 - LbRADH (mutant)
1zjy, 1zjz, 1zk0, 1zk1 – LbRADH (mutant) + NADH + alcohol
1zk4 - LbRADH (mutant) + NADH + acetophenone
Specific alcohol ADH
2cf5, 2cf6 – Cinnamyl-ADH – Arabidopsis thaliana
1piw, 1q1n, 1ps0 – Cinnamyl-yADH
3two - Cinnamyl-ADH + NADP – Helicobacter pylori
1m2w – Mannitol-ADH – Pseudomonas fluorescens
1w6s – Methanol-ADH – Methylobacterium extorquens
1yqx – Sinapyl-aADH II – aspen
1yqd – Sinapyl-aADH II + NADP
1bdb – Biphenyl dihydrodiol-ADH + NAD - Pseudomonas
Quinohemoprotein ADH
1kv9, 1yiq – PpQADH II + PQQ + heme – Pseudomonas putida
1kb0 - QADH I + PQQ + heme – Comamonas testosteroni
Hydroxyacyl-CoA dehydrogenase
Short chain HADH
1so8 – hSHCDH II – human
3rqs - hSHCDH
1f14 - hSHCDH (mutant)
Short chain HADH binary complex
1f12 - hSHCDH (mutant) + hydroxybutyryl-CoA
1f17, 1lsj, 1lso - hSHCDH (mutant) + NAD
1zbq - hSHCDH IV + NAD
1e3s - rSHCDH + NAD – rat
Short chain HADH ternary complex
1u7t - hSHCDH II + inhibitor + NAD
1f0y - hSHCDH + acetoacetyl-CoA + NAD
1il0, 1m75, 1m76 - hSHCDH (mutant) + acetoacetyl-CoA + NAD
1e3w - rSHCDH + 3-keto-butyrate + NAD
1e6w - rSHCDH + estradiol + NAD
Unspecified HADH
1uay - HADH II – Thermus thermophilus
1zej, 3ctv - HADH – Archaeoglobus fulgidus
1zcj - rHADH
2x58 - rHADH + CoA
2et6 – HADH (mutant) – Candida tropicalis
References
- ↑ Voet, et. al. Fundamentals of Biochemistry: 3rd Edition. Hoboken: Wiley & Sons, Inc, 2008.
- ↑ Protein: Alcohol Dehydrogenase. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm>
- ↑ Protein: Alcohol Dehydrogenase. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm>
- ↑ Protein: Alcohol Dehydrogenase. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm>
- ↑ Protein: Alcohol dehydrogenase from Human (Homo sapiens), different isozymes. SCOP. 2009. 1 March 2010 < http://scop.berkeley.edu/data/scop.b.d.c.b.b.c.html>
- ↑ Voet, et. al. Fundamentals of Biochemistry: 3rd Edition. Hoboken: Wiley & Sons, Inc, 2008.
- ↑ Protein: Alcohol Dehydrogenase. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm>
- ↑ Protein: Alcohol Dehydrogenase. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm>
- ↑ Voet, et. al. Fundamentals of Biochemistry: 3rd Edition. Hoboken: Wiley & Sons, Inc, 2008.
- ↑ Dickinson FM, Monger GP. A study of the kinetics and mechanism of yeast alcohol dehydrogenase with a variety of substrates. Biochem J. 1973 Feb;131(2):261-70. PMID:4352908
- ↑ Dickinson FM, Monger GP. A study of the kinetics and mechanism of yeast alcohol dehydrogenase with a variety of substrates. Biochem J. 1973 Feb;131(2):261-70. PMID:4352908
- ↑ Bille V, Remacle J. Simple-kinetic descriptions of alcohol dehydrogenase after immobilization on tresyl-chloride-activated agarose. Eur J Biochem. 1986 Oct 15;160(2):343-8. PMID:3769934
- ↑ Dickinson FM, Monger GP. A study of the kinetics and mechanism of yeast alcohol dehydrogenase with a variety of substrates. Biochem J. 1973 Feb;131(2):261-70. PMID:4352908
- ↑ Blomstrand R, Ostling-Wintzell H, Lof A, McMartin K, Tolf BR, Hedstrom KG. Pyrazoles as inhibitors of alcohol oxidation and as important tools in alcohol research: an approach to therapy against methanol poisoning. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3499-503. PMID:115004
- ↑ Alcohol Dehydrogenase. Worthington Biochemical Corporation . 31 March 2010 < http://http://www.worthington-biochem.com/ADH/default.html>
- ↑ Alcohol Dehydrogenase.Worthington Biochemical Corporation . 31 March 2010 < http://http://www.worthington-biochem.com/ADH/default.html>
- ↑ Goihberg E, Dym O, Tel-Or S, Levin I, Peretz M, Burstein Y. A single proline substitution is critical for the thermostabilization of Clostridium beijerinckii alcohol dehydrogenase. Proteins. 2007 Jan 1;66(1):196-204. PMID:17063493 doi:10.1002/prot.21170
- ↑ Goihberg E, Dym O, Tel-Or S, Shimon L, Frolow F, Peretz M, Burstein Y. Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution. Proteins. 2008 Feb 7;. PMID:18260103 doi:10.1002/prot.21946
- ↑ Goihberg E, Peretz M, Tel-Or S, Dym O, Shimon L, Frolow F, Burstein Y. Biochemical and Structural Properties of Chimeras Constructed by Exchange of Cofactor-Binding Domains in Alcohol Dehydrogenases from Thermophilic and Mesophilic Microorganisms. Biochemistry. 2010 Feb 9. PMID:20102159 doi:10.1021/bi901730x
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Joel L. Sussman, David Birrer