Insecticidal delta-endotoxin Cyt2Ba from Bacillus thuringiensis
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | <StructureSection load='' size='450' side='right' scene='Cyt2Ba/Cartoon_spectrum/2' caption=''> | ||
[[Image:Cyt_trim.jpg|left|150px]] | [[Image:Cyt_trim.jpg|left|150px]] | ||
| - | |||
| - | {{STRUCTURE_2rci| PDB=2rci | SCENE=Cyt2Ba/Cartoon_spectrum/2 }} | ||
| - | |||
| - | |||
===High-resolution crystal structure of activated Cyt2Ba monomer (δ-endotoxin) from ''Bacillus thuringiensis'' subsp. ''israelensis''=== | ===High-resolution crystal structure of activated Cyt2Ba monomer (δ-endotoxin) from ''Bacillus thuringiensis'' subsp. ''israelensis''=== | ||
| Line 9: | Line 6: | ||
{{Clear}} | {{Clear}} | ||
| - | |||
| - | <applet load='Cyt2Baa.pdb' size='500' frame='true' align='right' scene='Cyt2Ba/Cartoon_spectrum/2' /> | ||
| - | |||
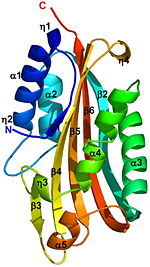
The [http://en.wikipedia.org/wiki/Crystal_structure crystal structure] of the [http://en.wikipedia.org/wiki/Proteolysis proteolytically] activated monomeric form of Cyt2Ba was determined at 1.8Å resolution. It consists of a single domain of <scene name='Cyt2Ba/Alpha_beta/5'>α/β</scene> architecture with a <scene name='Cyt2Ba/Beta/2'>β-sheet</scene> (yellow) surrounded by 2 <scene name='Cyt2Ba/Alpha/2'>α-helical</scene> layers <font color='red'><b>(red)</b></font> forming a cytolysin fold. The [http://en.wikipedia.org/wiki/Beta_sheet β-sheet] is comprised of 6 anti-parallel β-strands (β1-β6). On one side of this sheet there is an [http://en.wikipedia.org/wiki/Alpha_helix α-helix] layer consisting of α1, α2; and on the other side a second α-helix layer, composed of α3-α5. The β-strands β2-β5 of the central β-sheet have a modified Greek-key topology. The Greek key motif consists of four adjacent antiparallel strands and their linking loops. It consists of three antiparallel strands connected by hairpins, while the fourth is adjacent to the first and linked to the third by a longer loop [http://en.wikipedia.org/wiki/Beta_sheet]. <font color='gray'><b>Cyt2Ba (gray)</b></font> has only 16% sequence identity with <font color='red'><b>VVA2 (colored red,</b></font> [[1pp0]]), however they both have a cytolysin fold and their overall structure is very similar (see their <scene name='Cyt2Ba/Cyt2ba_vva/3'>structural alignment</scene>). | The [http://en.wikipedia.org/wiki/Crystal_structure crystal structure] of the [http://en.wikipedia.org/wiki/Proteolysis proteolytically] activated monomeric form of Cyt2Ba was determined at 1.8Å resolution. It consists of a single domain of <scene name='Cyt2Ba/Alpha_beta/5'>α/β</scene> architecture with a <scene name='Cyt2Ba/Beta/2'>β-sheet</scene> (yellow) surrounded by 2 <scene name='Cyt2Ba/Alpha/2'>α-helical</scene> layers <font color='red'><b>(red)</b></font> forming a cytolysin fold. The [http://en.wikipedia.org/wiki/Beta_sheet β-sheet] is comprised of 6 anti-parallel β-strands (β1-β6). On one side of this sheet there is an [http://en.wikipedia.org/wiki/Alpha_helix α-helix] layer consisting of α1, α2; and on the other side a second α-helix layer, composed of α3-α5. The β-strands β2-β5 of the central β-sheet have a modified Greek-key topology. The Greek key motif consists of four adjacent antiparallel strands and their linking loops. It consists of three antiparallel strands connected by hairpins, while the fourth is adjacent to the first and linked to the third by a longer loop [http://en.wikipedia.org/wiki/Beta_sheet]. <font color='gray'><b>Cyt2Ba (gray)</b></font> has only 16% sequence identity with <font color='red'><b>VVA2 (colored red,</b></font> [[1pp0]]), however they both have a cytolysin fold and their overall structure is very similar (see their <scene name='Cyt2Ba/Cyt2ba_vva/3'>structural alignment</scene>). | ||
| Line 20: | Line 14: | ||
The crystal structure of monomeric Cyt2Ba is the first structure of a [http://en.wikipedia.org/wiki/Toxicity toxic] form of the Cyt family. Its structure is [http://en.wikipedia.org/wiki/Homology_(biology) homologous] to the corresponding region of Cyt2Aa and to that of VVA2. This structural comparison shows that the toxicity of Cyt2Ba, Cyt2Aa and VVA2 is an inherent property of the monomer and not the result of secondary structure rearrangement upon cleavage. Solving the 3D structure of these proteins extends the knowledge of the cytolytic machinery of pore-forming toxins and helps in designing novel membrane-active cytotoxins. | The crystal structure of monomeric Cyt2Ba is the first structure of a [http://en.wikipedia.org/wiki/Toxicity toxic] form of the Cyt family. Its structure is [http://en.wikipedia.org/wiki/Homology_(biology) homologous] to the corresponding region of Cyt2Aa and to that of VVA2. This structural comparison shows that the toxicity of Cyt2Ba, Cyt2Aa and VVA2 is an inherent property of the monomer and not the result of secondary structure rearrangement upon cleavage. Solving the 3D structure of these proteins extends the knowledge of the cytolytic machinery of pore-forming toxins and helps in designing novel membrane-active cytotoxins. | ||
| + | </StructureSection> | ||
| + | __NOTOC__ | ||
==3D structures of δ-endotoxin== | ==3D structures of δ-endotoxin== | ||
Revision as of 07:38, 21 August 2013
| |||||||||||
3D structures of δ-endotoxin
Additional Resources
For additional information, see: Toxins
Reference
High-resolution crystal structure of activated Cyt2Ba monomer from Bacillus thuringiensis subsp. israelensis., Cohen S, Dym O, Albeck S, Ben-Dov E, Cahan R, Firer M, Zaritsky A, J Mol Biol. 2008 Jul 25;380(5):820-7. Epub 2008 May 11. PMID:18571667
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Joel L. Sussman, Eran Hodis, Jaime Prilusky, Michal Harel, David Canner, Eric Martz
Categories: Bacillus thuringiensis serovar israelensis | Single protein | Dym, O. | ISPC, Israel Structural Proteomics Center. | Alpha/beta architecture with beta-sheet surrounded by two alpha-helix layer | ISPC | Israel Structural Proteomics Center | Plasmid | Sporulation | Structural genomic | Toxin