Introduction
The single-stranded DNA-binding protein (SSB) of Escherichia coli is involved in all aspects of DNA metabolism: replication, repair, and recombination. In solution, the protein exists as a homotetramer of 18,843-kilodalton subunits. As it binds tightly and cooperatively to single-stranded DNA, it has become a prototypic model protein for studying protein-nucleic acid interactions. The sequences of the gene and protein are known, and the functional domains of subunit interaction, DNA binding, and protein-protein interactions have been probed by structure-function analyses of various mutations. The ssb gene has three promoters, one of which is inducible because it lies only two nucleotides from the LexA-binding site of the adjacent uvrA gene. Induction of the SOS response, however, does not lead to significant increases in SSB levels. The binding protein has several functions in DNA replication, including enhancement of helix destabilization by DNA helicases, prevention of reannealing of the single strands and protection from nuclease digestion, organization and stabilization of replication origins, primosome assembly, priming specificity, enhancement of replication fidelity, enhancement of polymerase processivity, and promotion of polymerase binding to the template. E. coli SSB is required for methyl-directed mismatch repair, induction of the SOS response, and recombinational repair. During recombination, SSB interacts with the RecBCD enzyme to find Chi sites, promotes binding of RecA protein, and promotes strand uptake.
Copies of SSB bind to the unwound DNA strands, keeping the strands separated so that both strands can serve as templates.
Structure
The structure of SSB consists of a homotetramer that has a DNA binding domain that binds to a single strand of DNA.
Binding Interactions in the Active Site
Residues involved in ssDNA binding
Spectroscopic studies also suggest that Trp 40 and Trp 54 form stacking interactions with the bases13, 14. Mutagenesis of Trp 40 and Trp 54 reduce ssDNA binding affinity14, 15. Crosslinking experiments16 and mutational studies17, 18 have shown that Phe 60 is involved in DNA binding. Consistent with these observations, Trp 40, Trp 54 and Phe 60, in the structure of the SSBc−ssDNA complex, make extensive interactions with the ssDNA (see above).
In the model of the SSBc−ssDNA structure presented here, Gly 15 is within 3.5 Å of the phosphate backbone at C4 (Fig. 2a). Mutation to Asp may sterically hinder ssDNA binding. The structure therefore suggests that G15D would affect ssDNA binding.
Finally, thermodynamic studies indicate that electrostatic interactions have a major role in SSB−ssDNA binding11, 20. The role of Lys residues and the N-terminus in ssDNA binding has also been probed by chemical modification21 and it was observed that acetylation of Lys 43, Lys 62, Lys 73, Lys 87, and the terminal amine is greatly reduced upon binding ssDNA. In the structure, these Lys residues, as well as the N-terminal amine, are within contact distance of the ssDNA backbone and selective acetylation of these residues would be expected to have a significant effect on ssDNA binding. Other basic residues make interactions with the ssDNA, either with the ssDNA bases (Arg 3) or with the phosphate backbone (Arg 84).
Overall, the chemical composition of the protein−ssDNA interface is mixed and a similar number of interactions are made between the protein and the bases or the protein and the ssDNA backbone.
The (SSB)65 binding mode
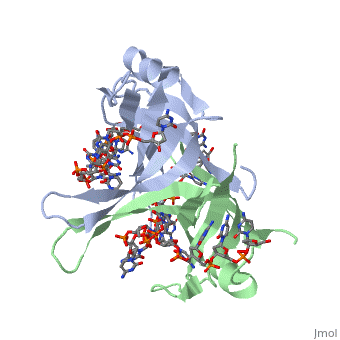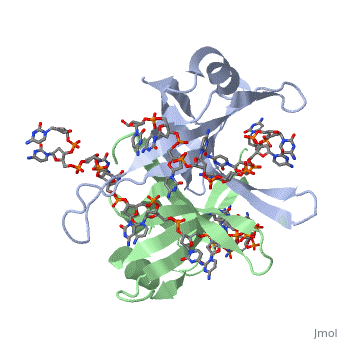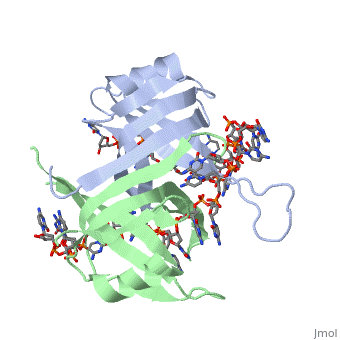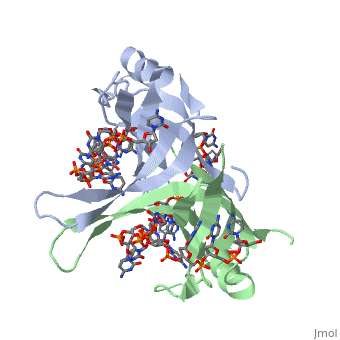
From the structure of SSBc bound to the two molecules of dC(pC)34, a model can be proposed for how a continuous stretch of ssDNA can interact with all four subunits and wrap around the tetramer. Such a model can be generated simply by applying the D2 symmetry operators (which relate the four subunits together) to the dC[pC]3−30 fragment to fill the gaps existing between the dC[pC]3−30, dC[pC]3−16, and dC[pC]19−27 ssDNA fragments. A model for SSBc bound to a long continuous ssDNA is presented in Fig. 4a. The proposed structural model for the (SSB)65 binding mode recapitulates remarkably well most of the biochemical and biophysical properties of this binding mode. First, the length of ssDNA occluded by the SSB tetramer is 65 nucleotides2. Second, ssDNA interacts with all four subunits, consistent with equilibrium fluorescence binding measurements2. Third, the ssDNA wraps around the outside of the tetramer7.
The (SSB)35 binding mode
A model for the (SSB)35 binding mode can also be postulated based on the structure of the SSBc−ssDNA complex by using the symmetry related complexes generated along the L45 loops. Note that the role of the L45 loop residues in intertetramer cooperativity has not been investigated in solution, and that evidence for the use of a tetramer−tetramer interface involving the L45 loops is uniquely crystallographic12 (also see above). Once the symmetry related SSBc−ssDNA complex structures that occur within the crystal are displayed, it becomes apparent that there is one path by which a long continuous stretch of ssDNA can interact with two adjacent SSBc tetramers to form a ssDNA bound filament of SSBc. This path is described in Fig. 4b,c. In this model, the ssDNA occupies the entire binding site of one SSBc subunit (indicated in cyan in Fig. 4b), half of the binding sites of two subunits (indicated in green and gold in Fig. 4b), and does not occupy the ssDNA binding site of the fourth subunit (in red in Fig. 4b). Although the (SSB)35 structural model is speculative, it is consistent with the biochemical information obtained for this binding mode. First, the occluded site size determined from the structural model is approx35 nucleotides per tetramer. Data from fluorescence equilibrium binding studies of SSB to oligodeoxynucleotides suggest that only two subunits of the SSB tetramer interact with ssDNA in the (SSB)35 mode2, 11. In fact, although the structural model in Fig. 4b indicates partial interactions with three of the subunits, the sum of these interactions is equivalent to those for only two subunits.
The structure of the SSBc−ssDNA complex reveals how the homotetrameric protein utilizes its perfectly symmetrical D2 structure to bind and compact a long stretch of ssDNA by wrapping it extensively around a relatively small protein. The structure also suggests a more speculative model by which the ssDNA can be compacted partially and still allow cooperative interactions between adjacent tetramers.
See Also
References
PMID: 2087220