Sandbox Reserved 828
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
---- | ---- | ||
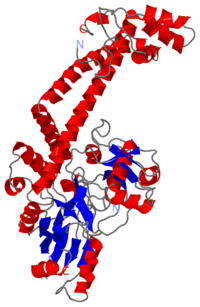
| - | <Structure load='1ab4' size='400' frame='true' align='right' caption='Breakage/Reunion domain at 2.8Å' scene='56/568026/Coloured/1'> | + | <Structure load='1ab4' size='400' frame='true' align='right' caption='Breakage/Reunion domain at 2.8Å (1ab4.pdb)' scene='56/568026/Coloured/1'> |
[[Image:1ab4.png|left|200px]] | [[Image:1ab4.png|left|200px]] | ||
| - | '''Introduction :''' | + | |
| + | =='''Introduction :'''== | ||
| + | |||
Gyrase is the '''only prokaryote DNA topoisomerase II able to introduce negative supercoils in the DNA''' in order to remove positive supercoils. It '''catalyses the hydrolysis of two phosphodiester bonds in a DNA segment''' (called G segment). Then, thanks to '''ATP dependant conformation changes''' it enables the passage of another segment (the T segment) through the break, and then religates the broken segment. Gyrase acts prior to the replication (before the replication fork) or other mecanisms requiring loose DNA. | Gyrase is the '''only prokaryote DNA topoisomerase II able to introduce negative supercoils in the DNA''' in order to remove positive supercoils. It '''catalyses the hydrolysis of two phosphodiester bonds in a DNA segment''' (called G segment). Then, thanks to '''ATP dependant conformation changes''' it enables the passage of another segment (the T segment) through the break, and then religates the broken segment. Gyrase acts prior to the replication (before the replication fork) or other mecanisms requiring loose DNA. | ||
| Line 15: | Line 17: | ||
---- | ---- | ||
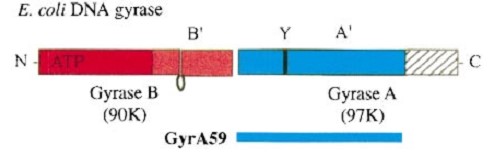
| - | '''Structure''' | + | =='''Structure'''== |
| - | + | ||
[[Image:Gyrasegene.jpg]] | [[Image:Gyrasegene.jpg]] | ||
| Line 39: | Line 40: | ||
---- | ---- | ||
| - | '''Reaction''' | + | =='''Reaction'''== |
| - | + | ||
The cleavage of DNA is achieved by a '''transesterification reaction between the tyrosines (Tyr122) and the target phosphoryl groups on opposing strands of the DNA backbone''', resulting in the tyrosine being covalentlty attached to the 5' end of the cleaved segment with a 4-base overhang. | The cleavage of DNA is achieved by a '''transesterification reaction between the tyrosines (Tyr122) and the target phosphoryl groups on opposing strands of the DNA backbone''', resulting in the tyrosine being covalentlty attached to the 5' end of the cleaved segment with a 4-base overhang. | ||
The '''active site tyrosines (Tyr 122)''' are on loops at either end of the dimer interface, 30Å apart, and sit at the ends of strongly basic grooves created by the dimer-related monomers. | The '''active site tyrosines (Tyr 122)''' are on loops at either end of the dimer interface, 30Å apart, and sit at the ends of strongly basic grooves created by the dimer-related monomers. | ||
The gyrase structure reveals a '''new cluster of conserved residues''', juxtaposing Tyr 122 and Arg 121 from one monomer and His 80, Arg 32 and Lys 42 from the other monomer. '''This cluster may form the active site of the breakage–reunion reaction''', with the other conserved positive charges (Arg 46 and Arg 47) anchoring the non-covalently bound 3' end of the cleaved DNA. | The gyrase structure reveals a '''new cluster of conserved residues''', juxtaposing Tyr 122 and Arg 121 from one monomer and His 80, Arg 32 and Lys 42 from the other monomer. '''This cluster may form the active site of the breakage–reunion reaction''', with the other conserved positive charges (Arg 46 and Arg 47) anchoring the non-covalently bound 3' end of the cleaved DNA. | ||
| + | <Structure load='1ab4_mm1.pdb' size='300' frame='true' align='right' caption='Biological Unit (dimer)' scene='56/568026/Biounit/1' /> | ||
Revision as of 16:10, 24 December 2013
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
| |||||||||||