Sandbox Reserved 828
From Proteopedia
(Difference between revisions)
| Line 37: | Line 37: | ||
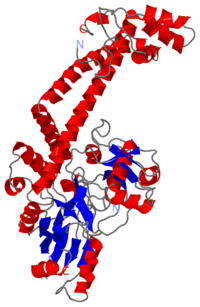
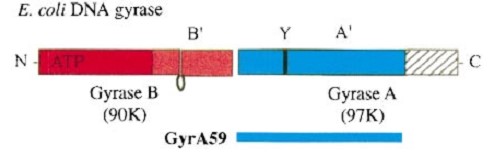
The remaining 30–35 kDa comprising the C-terminal domain (CTD) of GyrA shows a domain forming a b-pinwheel with a positively charged amino-acid perimeter. This carboxy-terminal domain of GyrA (cyan) is required for the introduction of DNA supercoils). | The remaining 30–35 kDa comprising the C-terminal domain (CTD) of GyrA shows a domain forming a b-pinwheel with a positively charged amino-acid perimeter. This carboxy-terminal domain of GyrA (cyan) is required for the introduction of DNA supercoils). | ||
| + | '''The B protein''' has the ATPase domain and the Toprim fold on it. Two ATPase domains dimerize to form a closed conformation. The Toprim fold is a Rossmann fold (beta-alpha-beta-alpha-beta) that contains three invariant acidic residues that coordinate magnesium ions involved in DNA cleavage and DNA religation. | ||
| - | + | ||
| - | The central core of the protein contains a Toprim fold and a DNA-binding core that contains a winged helix domain (WHD), often referred to as a CAP domain. The catalytic tyrosine lies on this WHD. The DNA-binding core consists of the WHD, which leads to a tower domain. . A coiled-coil region leads to a C-terminal domain that forms the main dimer interface | + | '''The central core''' of the protein contains a Toprim fold and a DNA-binding core that contains a winged helix domain (WHD), often referred to as a CAP domain. The catalytic tyrosine lies on this WHD. The DNA-binding core consists of the WHD, which leads to a tower domain. . A coiled-coil region leads to a C-terminal domain that forms the main dimer interface |
the Toprim domain is flexible and that this flexibility can allow the Toprim domain to coordinate with the WHD to form a competent cleavage complex | the Toprim domain is flexible and that this flexibility can allow the Toprim domain to coordinate with the WHD to form a competent cleavage complex | ||
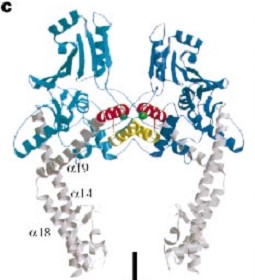
DNA was bent by ~150 degrees through an invariant isoleucine (in topoisomerase II it is I833 and in gyrase it is I172) | DNA was bent by ~150 degrees through an invariant isoleucine (in topoisomerase II it is I833 and in gyrase it is I172) | ||
| Line 45: | Line 46: | ||
The structures formed a novel beta barrel, which bends DNA by wrapping the nucleic acid around itself. The bending of DNA by gyrase has been proposed as a key mechanism in the ability of gyrase to introduce negative supercoils into the DNA. This is consistent with footprinting data that shows | The structures formed a novel beta barrel, which bends DNA by wrapping the nucleic acid around itself. The bending of DNA by gyrase has been proposed as a key mechanism in the ability of gyrase to introduce negative supercoils into the DNA. This is consistent with footprinting data that shows | ||
that gyrase has a 140-base-pair footprint. Both gyrase and topoisomerase IV CTDs bend DNA, but only gyrase introduces negative supercoils. | that gyrase has a 140-base-pair footprint. Both gyrase and topoisomerase IV CTDs bend DNA, but only gyrase introduces negative supercoils. | ||
| + | |||
| + | |||
| + | ---- | ||
| + | '''Mecanism''' | ||
| + | |||
| + | 40-bp of duplex DNA, the G-segment, bind to the core of the enzyme and are cleaved by the active site tyrosines, while another DNA duplex, the T-segment, is captured through the ATP-induced dimerization of a protein gate, the N-gate. After passage through the transiently broken G-segment (DNA gate), the T-segment exits the protein through another protein gate, the C-gate. ATP hydrolysis and release reset the conformation of the enzyme and DNA to their initial state, poised for another strand-passage event or release of the DNA. | ||
| + | N- and C-gates that open or close to allow T-segment transport through both theprotein and the cleaved G-segment. | ||
| + | |||
---- | ---- | ||
Revision as of 16:20, 24 December 2013
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
| |||||||||||