TAL effector
From Proteopedia
(Difference between revisions)
m (Sandbox Reserved 701 moved to TAL effector: requested by Editor) |
|||
| Line 1: | Line 1: | ||
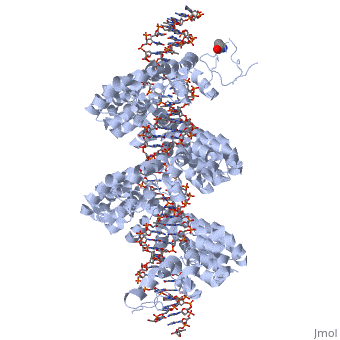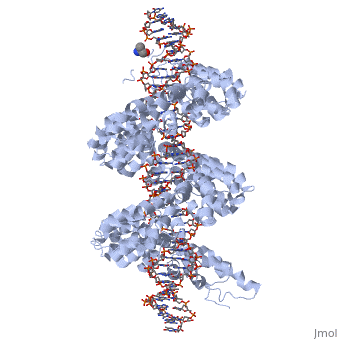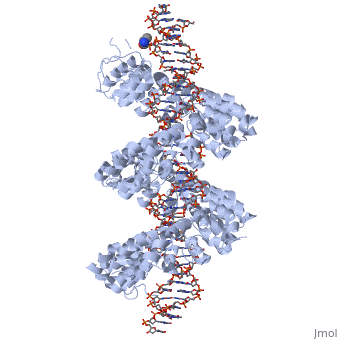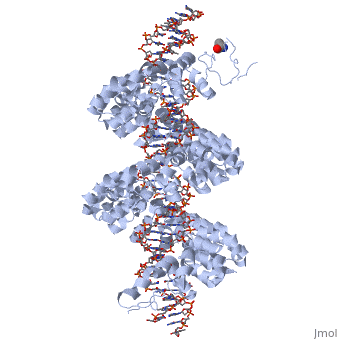
| - | < | + | <StructureSection load='3ugm' size='350' side='right' caption='Structure of TAL effector PthXo1 bound to its target DNA [[3ugm]]' scene=''> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
'''Transcription activator-like effector PthXo1''' (TALE PthXo1) TALEs are proteins originally derived from the plant pathogene Xanthomonas spp. The gene encoding for the effector are Pthxo1 and Pxo_00227. Originally they were detected in [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2432079/ Xanthomonas Oryzae] but the TALE PthXo1 genes were transformed into [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3100984/ Escherichia Coli] to express the DNA-binding protein. | '''Transcription activator-like effector PthXo1''' (TALE PthXo1) TALEs are proteins originally derived from the plant pathogene Xanthomonas spp. The gene encoding for the effector are Pthxo1 and Pxo_00227. Originally they were detected in [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2432079/ Xanthomonas Oryzae] but the TALE PthXo1 genes were transformed into [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3100984/ Escherichia Coli] to express the DNA-binding protein. | ||
| - | |||
| - | <!--<scene name='Sandbox_Reserved_701/Dna/1'>DNA</scene> | ||
| - | <scene name='Sandbox_Reserved_701/Tale/5'>Tale</scene>--> | ||
| - | |||
{{ABSTRACT_PUBMED_022223736}} <ref> PMID:022223736 </ref> | {{ABSTRACT_PUBMED_022223736}} <ref> PMID:022223736 </ref> | ||
The interaction between the <scene name='Sandbox_Reserved_701/Tale/5'>TAL effector PthXo1</scene> and a <scene name='Sandbox_Reserved_701/Dna/1'>DNA double helix</scene> is displayed on the right side. | The interaction between the <scene name='Sandbox_Reserved_701/Tale/5'>TAL effector PthXo1</scene> and a <scene name='Sandbox_Reserved_701/Dna/1'>DNA double helix</scene> is displayed on the right side. | ||
| - | <!--==TEST== | ||
| - | |||
| - | {{STRUCTURE_4gg4| PDB=4gg4 | SCENE= }} | ||
| - | |||
| - | {{STRUCTURE_4gg4| PDB=4gg4 | SIZE=500|left|CAPTION=Structure of another TAL effector bound to its target DNA [[4gg4]] }}--> | ||
| - | |||
| - | |||
| - | |||
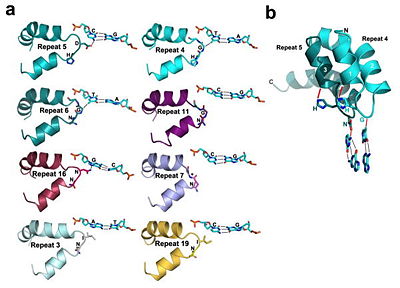
<br>[[Image:overview.jpg|left|300px|thumb|Fig.1 Structure of the PthXo1 DNA binding region in complex with its target site. The coloring of individual repeats matches the schematic in XXX]]<br> | <br>[[Image:overview.jpg|left|300px|thumb|Fig.1 Structure of the PthXo1 DNA binding region in complex with its target site. The coloring of individual repeats matches the schematic in XXX]]<br> | ||
| - | |||
| - | ==3D structures of TAL effectors== | ||
| - | |||
| - | [[3ugm]] - XoTAL pthxo1 + DNA - ''Xanthomonas oryzae''<br /> | ||
| - | [[3v6p]] - XoTAL dHax3 <br /> | ||
| - | [[4gg4]] – XoTAL dHax3 + DNA + RNA<br /> | ||
| - | [[3v6t]] – XoTAL dHax3 + DNA <br /> | ||
| - | [[4gjr]], [[4gjp]] – XoTAL dHax3 (mutant) + DNA<br /> | ||
| - | |||
| - | |||
| - | [[Category:Topic Page]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | =='''About this Structure'''== | ||
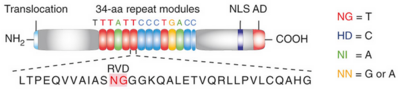
===DNA binding module with marked RVD=== | ===DNA binding module with marked RVD=== | ||
| - | |||
| - | <!--[[Image:Positions_of_the_mutations_in_PR_variants_used_for_structural_studies.jpg|left|320px|thumb| Fig.3 Positions of the mutations in PR variants used for structural studies. --> | ||
| - | |||
| - | |||
| - | |||
<br>[[Image:sanjana.png|left|400px|thumb|Fig.2]]<br> | <br>[[Image:sanjana.png|left|400px|thumb|Fig.2]]<br> | ||
'''Figure 2''': Natural structure of TALEs derived from Xanthomonas sp. Each DNA-binding module consists of 34 amino acids, where the RVDs in the 12th and 13th amino acid positions of each repeat specify the DNA base being targeted according to the cipher NG = <scene name='Sandbox_Reserved_701/T_blau/1'>T</scene>, HD = <scene name='Sandbox_Reserved_701/C_schwarz/1'>C</scene>, NI = <scene name='Sandbox_Reserved_701/A_gruen/1'>A</scene>, and NN = <scene name='Sandbox_Reserved_701/G_gelb/1'>G</scene> or <scene name='Sandbox_Reserved_701/A_gruen/1'>A</scene>. The DNA-binding modules are flanked by nonrepetitive N and C termini, which carry the translocation, nuclear localization (NLS) and transcription activation (AD) domains. A cryptic signal within the N terminus specifies a thymine as the first base of the target site.<ref> PMID:022223736 </ref> | '''Figure 2''': Natural structure of TALEs derived from Xanthomonas sp. Each DNA-binding module consists of 34 amino acids, where the RVDs in the 12th and 13th amino acid positions of each repeat specify the DNA base being targeted according to the cipher NG = <scene name='Sandbox_Reserved_701/T_blau/1'>T</scene>, HD = <scene name='Sandbox_Reserved_701/C_schwarz/1'>C</scene>, NI = <scene name='Sandbox_Reserved_701/A_gruen/1'>A</scene>, and NN = <scene name='Sandbox_Reserved_701/G_gelb/1'>G</scene> or <scene name='Sandbox_Reserved_701/A_gruen/1'>A</scene>. The DNA-binding modules are flanked by nonrepetitive N and C termini, which carry the translocation, nuclear localization (NLS) and transcription activation (AD) domains. A cryptic signal within the N terminus specifies a thymine as the first base of the target site.<ref> PMID:022223736 </ref> | ||
| - | |||
| - | |||
| - | |||
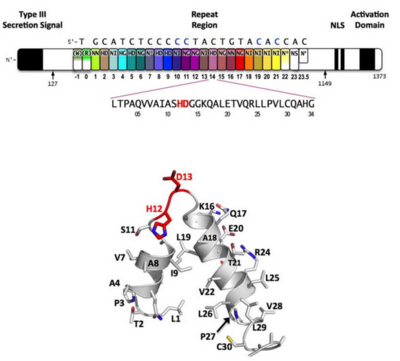
===Tertiary structure of the RVD=== | ===Tertiary structure of the RVD=== | ||
| Line 78: | Line 22: | ||
Sequence-specific contacts of PthXo1 to the DNA are made exclusively by the second residue in each RVD to the corresponding base on the sense strand. In contrast, the side chain at the first position of each RVD contacts the backbone carbonyl oxygen of position 8 in each repeat, constraining the RVD-containing loop (Figure 3). Additional, nonspecific contacts to the DNA are made by a lysine and glutamine found at positions 16 and 17. | Sequence-specific contacts of PthXo1 to the DNA are made exclusively by the second residue in each RVD to the corresponding base on the sense strand. In contrast, the side chain at the first position of each RVD contacts the backbone carbonyl oxygen of position 8 in each repeat, constraining the RVD-containing loop (Figure 3). Additional, nonspecific contacts to the DNA are made by a lysine and glutamine found at positions 16 and 17. | ||
| - | |||
| - | |||
| - | |||
| - | |||
===Interaction between RVDs and DNA=== | ===Interaction between RVDs and DNA=== | ||
| - | |||
| - | |||
‘HD’ RVDs: | ‘HD’ RVDs: | ||
the aspartate residue makes [http://en.wikipedia.org/wiki/Van_der_Waals_force van der Waals] contacts with the edge of the corresponding cytosine base and a [http://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bond] to the cytosine N4 atom. | the aspartate residue makes [http://en.wikipedia.org/wiki/Van_der_Waals_force van der Waals] contacts with the edge of the corresponding cytosine base and a [http://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bond] to the cytosine N4 atom. | ||
| - | |||
‘NG’ and ‘HG’ RVDs: | ‘NG’ and ‘HG’ RVDs: | ||
make a contact in which the backbone alpha carbon of the glycine residue forms a nonpolar van der Waals interaction with the methyl group of the opposing thymine base (average distance ~ 3.3 Å). At the one position where an NG is aligned opposite a cytosine base, the backbone carbonyl and alpha-carbon of the same glycine residue displays a less favorable, far more distant contact (~ 6 Å). | make a contact in which the backbone alpha carbon of the glycine residue forms a nonpolar van der Waals interaction with the methyl group of the opposing thymine base (average distance ~ 3.3 Å). At the one position where an NG is aligned opposite a cytosine base, the backbone carbonyl and alpha-carbon of the same glycine residue displays a less favorable, far more distant contact (~ 6 Å). | ||
| - | |||
‘NN’ RVDs: | ‘NN’ RVDs: | ||
| Line 104: | Line 40: | ||
PthXo1 contains two 33 residue (7 and 22). Since RVDs are followed immediately by two conserved glycine residues, this repeat is equivalent to an ‘NG’ repeat in which one of those glycine residues is missing. The crystal structure indicates that the deletion results in a truncated RVD loop that extends less deeply into the DNA major groove, with the glycine at position 13 located a considerable distance (over 6 Å) | PthXo1 contains two 33 residue (7 and 22). Since RVDs are followed immediately by two conserved glycine residues, this repeat is equivalent to an ‘NG’ repeat in which one of those glycine residues is missing. The crystal structure indicates that the deletion results in a truncated RVD loop that extends less deeply into the DNA major groove, with the glycine at position 13 located a considerable distance (over 6 Å) | ||
| - | |||
‘NI’ RVDs: | ‘NI’ RVDs: | ||
Occurs seven times in PthXo1, and displays an unusual contact pattern to adenosine or cytosine bases. The aliphatic side chain of the isoleucine residue is observed to make non-polar van der Waals contacts to C8 (and N7) of the adenine purine ring, or to C5 of the cytosine pyrimidine ring. | Occurs seven times in PthXo1, and displays an unusual contact pattern to adenosine or cytosine bases. The aliphatic side chain of the isoleucine residue is observed to make non-polar van der Waals contacts to C8 (and N7) of the adenine purine ring, or to C5 of the cytosine pyrimidine ring. | ||
| - | |||
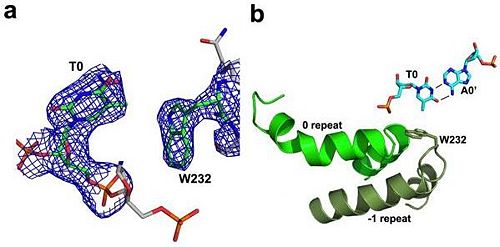
N-terminal to the canonical repeats, the PthXo1 structure reveals two degenerate repeat folds that appear to cooperate to specify the conserved thymine that precedes the RVD-specified sequence (Figure 5). We have designated these as the 0th and -1st repeats. Residues 221 to 239 and residues 256 to 273 each form a helix and an adjoining loop that resembles helix 1 and the RVD loop in the canonical repeats; the remaining residues in each region are poorly ordered. Those two N-terminal regions converge near the 5′ thymine base, with the indole ring of tryptophan 232 (in the -1st repeat) making a van der Waals contact with the methyl group of that base. Mutation of the thymine reduces TAL effector activity at the target (3, 15). Tryptophan 232, as well as the surrounding residues, is highly conserved across available, intact TAL effector sequences. | N-terminal to the canonical repeats, the PthXo1 structure reveals two degenerate repeat folds that appear to cooperate to specify the conserved thymine that precedes the RVD-specified sequence (Figure 5). We have designated these as the 0th and -1st repeats. Residues 221 to 239 and residues 256 to 273 each form a helix and an adjoining loop that resembles helix 1 and the RVD loop in the canonical repeats; the remaining residues in each region are poorly ordered. Those two N-terminal regions converge near the 5′ thymine base, with the indole ring of tryptophan 232 (in the -1st repeat) making a van der Waals contact with the methyl group of that base. Mutation of the thymine reduces TAL effector activity at the target (3, 15). Tryptophan 232, as well as the surrounding residues, is highly conserved across available, intact TAL effector sequences. | ||
| + | </StructureSection> | ||
| + | ==3D structures of TAL effectors== | ||
| + | [[3ugm]] - XoTAL pthxo1 + DNA - ''Xanthomonas oryzae''<br /> | ||
| + | [[3v6p]] - XoTAL dHax3 <br /> | ||
| + | [[4gg4]] – XoTAL dHax3 + DNA + RNA<br /> | ||
| + | [[3v6t]] – XoTAL dHax3 + DNA <br /> | ||
| + | [[4gjr]], [[4gjp]] – XoTAL dHax3 (mutant) + DNA<br /> | ||
==Reference== | ==Reference== | ||
| Line 123: | Line 64: | ||
==Contributors== | ==Contributors== | ||
Fabian Stritt, Philipp Warmer | Fabian Stritt, Philipp Warmer | ||
| - | + | [[Category:Topic Page]] | |
| - | + | ||
[[Category: Xanthomonas oryzae]] | [[Category: Xanthomonas oryzae]] | ||
[[Category: Bogdanove, A J.]] | [[Category: Bogdanove, A J.]] | ||
Revision as of 17:16, 9 February 2014
| |||||||||||
3D structures of TAL effectors
3ugm - XoTAL pthxo1 + DNA - Xanthomonas oryzae
3v6p - XoTAL dHax3
4gg4 – XoTAL dHax3 + DNA + RNA
3v6t – XoTAL dHax3 + DNA
4gjr, 4gjp – XoTAL dHax3 (mutant) + DNA
Reference
- Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The Crystal Structure of TAL Effector PthXo1 Bound to Its DNA Target. Science. 2012 Jan 5. PMID:22223736 doi:10.1126/science.1216211
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012 Jan 5;7(1):171-92. doi: 10.1038/nprot.2011.431. PMID:22222791 doi:10.1038/nprot.2011.431
- Barberousse V, Sacco D, Dellacherie E. Chromatographic evaluation of the binding of haemoglobin to polyanionic polymers. J Chromatogr. 1986 Nov 7;369(1):244-7. PMID:2432079
Contributors
Fabian Stritt, Philipp Warmer
Proteopedia Page Contributors and Editors (what is this?)
Philipp Warmer, Michal Harel, Jaime Prilusky, OCA, Alexander Berchansky