User:Cody Couperus/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 27: | Line 27: | ||
Prothrombin is activated by prothrombinase which consists of FXa, FVa, calcium, and a phospholipid surface. In vivo the first cleavage occurs at the R320-I321 bond, corresponding to residues 15-16 in thrombin which is the N-terminus of the B chain, producing meizothrombin.<ref>PMID: 22944689</ref> Subsequent cleavage at R271-T272 yields thrombin.<ref>PMID: 22944689</ref> The initial cleavage can also occur at R271 resulting in prothrombin-2 which will then be cleaved at R320 to produce thrombin.<ref>PMID: 1995649</ref> | Prothrombin is activated by prothrombinase which consists of FXa, FVa, calcium, and a phospholipid surface. In vivo the first cleavage occurs at the R320-I321 bond, corresponding to residues 15-16 in thrombin which is the N-terminus of the B chain, producing meizothrombin.<ref>PMID: 22944689</ref> Subsequent cleavage at R271-T272 yields thrombin.<ref>PMID: 22944689</ref> The initial cleavage can also occur at R271 resulting in prothrombin-2 which will then be cleaved at R320 to produce thrombin.<ref>PMID: 1995649</ref> | ||
| - | After cleavage by prothrombinase the new B chain N-terminus (Ile16) folds into the core protease domain and forms a salt bridge with Asp194.<ref>PMID: 22944689</ref> This leads to stabilization of regions of the 180s-loop, Na+ binding loop, and γ-loop (zymogen activation domains). These changes provide the correct conformation for the S1 pocket and | + | After cleavage by prothrombinase the new B chain N-terminus (Ile16) folds into the core protease domain and forms a salt bridge with Asp194.<ref>PMID: 22944689</ref> This leads to stabilization of regions of the 180s-loop, Na+ binding loop, and γ-loop (zymogen activation domains). These changes provide the correct conformation for the S1 pocket and oxyanion hole for catalysis.<ref>PMID: 22944689</ref><ref>PMID: 15890651</ref> |
| Line 43: | Line 43: | ||
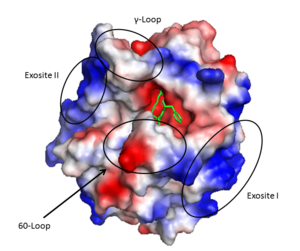
The <scene name='58/583418/B_chain/1' target='0'>B chain</scene> contains the active site of the protein and has numerous notable structural features. The active site is formed at the rims of two interacting 6 stranded <scene name='58/583418/Beta_barrel/1'>beta barrel domains</scene> which are surrounded by 4 helical regions and many turns. | The <scene name='58/583418/B_chain/1' target='0'>B chain</scene> contains the active site of the protein and has numerous notable structural features. The active site is formed at the rims of two interacting 6 stranded <scene name='58/583418/Beta_barrel/1'>beta barrel domains</scene> which are surrounded by 4 helical regions and many turns. | ||
| - | The serine protease <scene name='58/583418/Catalytic_triad/1'>catalytic triad</scene>, based on chymotrypsin numbering, are Ser195, His57, and Asp102. As is common with serine proteases, an | + | The serine protease <scene name='58/583418/Catalytic_triad/1'>catalytic triad</scene>, based on chymotrypsin numbering, are Ser195, His57, and Asp102. As is common with serine proteases, an <scene name='58/583418/Oxyanion_hole/2'>oxyanion hole</scene> hole is formed by backbone amides of Ser195 and Gly193.<ref>PMID: 22944689</ref> This has the functional role of stabilizing the oxyanion intermediate involved in the serine protease mechanism by hydrogen bonding to the oxygen of the P1 residue (traditional substrate-protein nomeclature <ref>PMID: 22925665</ref>. In addition, since thrombin cleaves after Arg/Lys the <scene name='58/583418/S1/1'>S1 specificity site</scene>, formed by the 180s- and 220s- loops, has Asp189 at the base to form a salt bridge with the incoming substrate. Furthermore, the S4 binding pocket accommodates hydrophobic substrate residues. |
The '''active site''' cleft rims are formed by the hydrophobic and rigid <scene name='58/583418/Loops/1'>60-loops</scene> (residues L60, Y60a, P60b, P60c, W60d, D60e, K60f, N60g, F60h, T60i, and N60g) and the <scene name='58/583418/Loops/1'>γ-loop</scene> (residues T147, W147a, T147b, A147c, N147d, and V147f) while the base is mostly hydrophilic negatively charged amino acids. The cleft is deep compared to more promiscuous serine proteases, consequently substrates must either have a large loop that is cleaved or have favorable interactions with the insertion loops <ref>PMID: 16102053</ref>. | The '''active site''' cleft rims are formed by the hydrophobic and rigid <scene name='58/583418/Loops/1'>60-loops</scene> (residues L60, Y60a, P60b, P60c, W60d, D60e, K60f, N60g, F60h, T60i, and N60g) and the <scene name='58/583418/Loops/1'>γ-loop</scene> (residues T147, W147a, T147b, A147c, N147d, and V147f) while the base is mostly hydrophilic negatively charged amino acids. The cleft is deep compared to more promiscuous serine proteases, consequently substrates must either have a large loop that is cleaved or have favorable interactions with the insertion loops <ref>PMID: 16102053</ref>. | ||
| Line 53: | Line 53: | ||
'''Exosite II''' is also part of the B chain, and derived from numerous basic amino acids, this is the site of heparin binding through the sulfate groups on the glycosaminoglycan. It is also the site of GpIα on the platelet surface. | '''Exosite II''' is also part of the B chain, and derived from numerous basic amino acids, this is the site of heparin binding through the sulfate groups on the glycosaminoglycan. It is also the site of GpIα on the platelet surface. | ||
| - | The <scene name='58/583418/Sodium_binding_loop/1'>sodium binding site</scene> is formed by the 180s- and 220s- loops. Na+ is bound by the backbone oxygens of Arg221a and Lys224 in addition to four water molecules in a classic octahedral geometry<ref>PMID: 9108691</ref>. Through the covelent disulfide linkage between Cys220 and Cys 191 the sodium binding site is linked to Ser195 and the | + | The <scene name='58/583418/Sodium_binding_loop/1'>sodium binding site</scene> is formed by the 180s- and 220s- loops. Na+ is bound by the backbone oxygens of Arg221a and Lys224 in addition to four water molecules in a classic octahedral geometry<ref>PMID: 9108691</ref>. Through the covelent disulfide linkage between Cys220 and Cys 191 the sodium binding site is linked to Ser195 and the oxyanion hole. |
==Thrombin Allostery== | ==Thrombin Allostery== | ||
| - | Binding of thrombin by sodium or at exosite I stabilizes a form of thrombin that improves substrate recognition.<ref>PMID: 22944689</ref> This occurs due to energetic linkage between these sites to the S1 binding pocket and | + | Binding of thrombin by sodium or at exosite I stabilizes a form of thrombin that improves substrate recognition.<ref>PMID: 22944689</ref> This occurs due to energetic linkage between these sites to the S1 binding pocket and oxyanion hole. Rapid kinetic analysis suggests that thrombin is in a dynamic equilibrium that consists of a fast, slow, and inactive state<ref>PMID: 22944689</ref>. There is question as to the physiologic relevance of the inactive state. Regardless, there will be a proportion of fast:slow thrombin and sodium binding to the fast form stabilizes that conformation. Indeed, mutation of the residues involved in sodium binding diminishes the activity of thrombin.<ref>PMID: 22944689</ref> It should be restated, that current data suggest that sodium binding does not induce a conformation change, rather, it stabilizes a conformation of thrombin that has greater activity. |
Revision as of 01:02, 30 April 2014
Thrombin: Structure and Function
| |||||||||||