Sandbox Reserved 933
From Proteopedia
| Line 8: | Line 8: | ||
== Introduction == | == Introduction == | ||
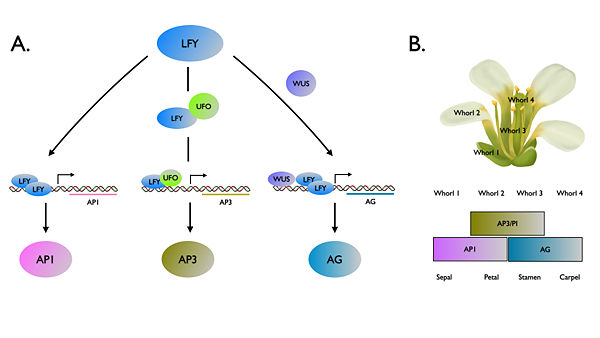
[[Image:Organ_identity.jpg|600px|left|thumb| Figure 1. Schematic presentation LEAFY regulatory roles in controlling floral organ identity (A) and the ABC model in ''Arabidopsis thaliana'' (B). ]] | [[Image:Organ_identity.jpg|600px|left|thumb| Figure 1. Schematic presentation LEAFY regulatory roles in controlling floral organ identity (A) and the ABC model in ''Arabidopsis thaliana'' (B). ]] | ||
| - | '''''FLORICAULA/LEAFY'' (''FLO/LFY'')''' genes encode a plant specific transcription factor family that controlling floral fate of reproductive phase. <ref name="Weigel1992"> Detlef Weigel, John Alvarez, David R. Smyth, Martin F. Yanofsky, Elliot M. Meyerowitz, LEAFY controls floral meristem identity in Arabidopsis. Cell 69 :843-859, http://dx.doi.org/10.1016/0092-8674(92)90295-N.</ref> In the plant model system ''Arabidopsis thaliana '', ‘’LFY’’ also acts upstream of floral homeotic genes to modulate organ identity. <ref name= ''Irish2010'' > Irish, V. F. (2010), The flowering of Arabidopsis flower development. The Plant Journal, 61: 1014–1028. http://dx.doi.org/10.1111/j.1365-313X.2009.04065.x </ref> LFY activates the organ identity genes by binding to promoter regions of floral organ identity genes. LFY can directly bind to the promoter to '''''APELATA1'' (''AP1'')''', while co-regulators '''''UNUSUAL FLORAL ORGANS'' (''UFO'')''' <ref name= ''Chae2008'' >Chae, E., Tan, Q.K., Hill, T.A. & Irish, V.F. 2008. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135:1235-45 http://dx.doi.org/10.1242/dev.015842</ref> and '''''WUSCHEL'' (''WUS'')'''<ref name= ''Hong2003'' >HONG, R.L., HAMAGUCHI, L., BUSCH, M.A. and WEIGEL, D. (2003). Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15: 1296-1309. http://dx.doi.org/10.1105/tpc.009548</ref> are required for increment of binding affinity to promoter regions of '''''APELATA3'' (''AP3'')''' and '''''AGAMOUS'' (''AG'')''', respectively. The exact mechanism how LFY binds to these promoters has yet to be well elucidated until the first structure report about <scene name='57/579703/Ap1-dimer/1'>LFY-pAP1</scene> and <scene name='57/579703/2vy2_assembly/1'>LFY-pAG</scene><ref name= ''Hames2008'' >Hames, C., Ptchelkine, D., Grimm, C., Thevenon, E., Moyroud, E., Gérard, F. Martiel, J.L., Benlloch, R., Parcy, F. & Müller, C.W. 2008. Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO Journal 27:2628-2637. http://dx.doi.org/10.1038/emboj.2008.184 | + | '''''FLORICAULA/LEAFY'' (''FLO/LFY'')''' genes encode a plant specific transcription factor family that controlling floral fate of reproductive phase. <ref name="Weigel1992"> Detlef Weigel, John Alvarez, David R. Smyth, Martin F. Yanofsky, Elliot M. Meyerowitz, LEAFY controls floral meristem identity in Arabidopsis. Cell 69 :843-859, http://dx.doi.org/10.1016/0092-8674(92)90295-N.</ref><ref name="Coen1990"> Coen, E.S., Romero, J.M., Doyle, S., Elliot, R., Murphy, G. & Carpenter, R. (1990) ''floricaula'': a homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 http://dx.doi.org/10.1016/0092-8674(90)90426-F</ref>. In the plant model system ''Arabidopsis thaliana '', ‘’LFY’’ also acts upstream of floral homeotic genes to modulate organ identity. <ref name= ''Irish2010'' > Irish, V. F. (2010), The flowering of Arabidopsis flower development. The Plant Journal, 61: 1014–1028. http://dx.doi.org/10.1111/j.1365-313X.2009.04065.x </ref> LFY activates the organ identity genes by binding to promoter regions of floral organ identity genes. LFY can directly bind to the promoter to '''''APELATA1'' (''AP1'')''', while co-regulators '''''UNUSUAL FLORAL ORGANS'' (''UFO'')''' <ref name= ''Chae2008'' >Chae, E., Tan, Q.K., Hill, T.A. & Irish, V.F. 2008. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135:1235-45 http://dx.doi.org/10.1242/dev.015842</ref> and '''''WUSCHEL'' (''WUS'')'''<ref name= ''Hong2003'' >HONG, R.L., HAMAGUCHI, L., BUSCH, M.A. and WEIGEL, D. (2003). Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15: 1296-1309. http://dx.doi.org/10.1105/tpc.009548</ref> are required for increment of binding affinity to promoter regions of '''''APELATA3'' (''AP3'')''' and '''''AGAMOUS'' (''AG'')''', respectively. The exact mechanism how LFY binds to these promoters has yet to be well elucidated until the first structure report about <scene name='57/579703/Ap1-dimer/1'>LFY-pAP1</scene> and <scene name='57/579703/2vy2_assembly/1'>LFY-pAG</scene><ref name= ''Hames2008'' >Hames, C., Ptchelkine, D., Grimm, C., Thevenon, E., Moyroud, E., Gérard, F. Martiel, J.L., Benlloch, R., Parcy, F. & Müller, C.W. 2008. Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO Journal 27:2628-2637. http://dx.doi.org/10.1038/emboj.2008.184 |
| - | </ref> . Among land plants, FLO/LFY homologs share a highly conserved DNA binding region that a hypothesis claimed substitution in this domain might result in the functional divergence<ref name= ''Maize2005'' >MAIZEL, A., BUSCH, M.A., TANAHASHI, T., PERKOVIC, J., KATAO, M., HASEBE, M. and WEIGEL, D. (2005). The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308: 260-263. http://dx.doi.org/10.1126/science.1108229</ref> . Recently, a new study | + | </ref> . Among land plants, FLO/LFY homologs share a highly conserved DNA binding region that a hypothesis claimed substitution in this domain might result in the functional divergence<ref name= ''Maize2005'' >MAIZEL, A., BUSCH, M.A., TANAHASHI, T., PERKOVIC, J., KATAO, M., HASEBE, M. and WEIGEL, D. (2005). The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308: 260-263. http://dx.doi.org/10.1126/science.1108229</ref> . Recently, a new study provided new insights of structural basis of LEAFY evolution by changing DNA binding activity<ref name= ''Sayou2014'' >Sayou, C., Monniaux, M., Nanao, M.H., Moyroud, E., Brockington, S.F., Thévenon, E., Chahtane, H., Warthmann, N., Melkonian, M., Zhang, Y., Wong, G., Weigel, D., Parcy, F. and Dumas, R. 2014. A Promiscuous Intermediate Underlies the Evolution of LEAFY DNA Binding Specificity Science 343: 645-648 http://dx.doi.org/10.1126/science.1248229</ref>. |
| Line 16: | Line 16: | ||
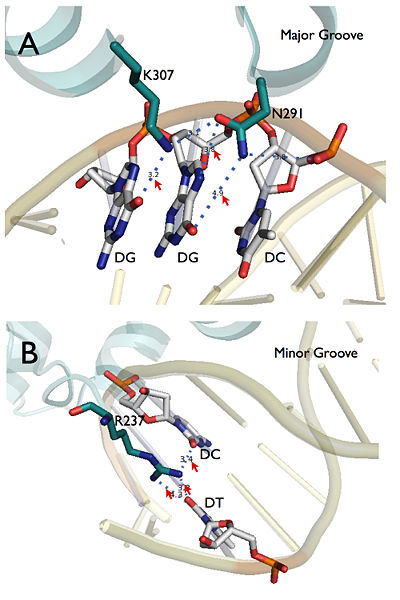
[[Image:DNA_Recognition.jpg|400px|left|thumb| Figure 2. Site specific recognition of LFY protein at major (A) and minor (B) grooves. Assembly of PDB entry 2VY1 were obtained from PISA server and further visualized by Pymol. Red arrows marked site specific hydrogen bonds.]] | [[Image:DNA_Recognition.jpg|400px|left|thumb| Figure 2. Site specific recognition of LFY protein at major (A) and minor (B) grooves. Assembly of PDB entry 2VY1 were obtained from PISA server and further visualized by Pymol. Red arrows marked site specific hydrogen bonds.]] | ||
=== General information about the structure === | === General information about the structure === | ||
| - | The ''LFY'' gene encodes a 424 amino acids protein that containing two domains. The N-terminal domain of LFY has been proved mediating homodimerization (ref) and it is also thought to be responsible for transcriptional activation | + | The ''LFY'' gene encodes a 424 amino acids protein that containing two domains. The N-terminal domain of LFY has been proved mediating homodimerization (ref) and it is also thought to be responsible for transcriptional activation <ref name="Hames et al." />. The C-terminal consensus is highly conserved among land species and functioning as DNA-binding domain. Two DNA-protein binding structure for LEAFY were first published by Hame et al. 2008 <ref name="Weigel1992" /><ref name="Coen1990" />. These two structures include a recombinant C-terminal domain of LEAFY expressed by ''Escherichia coli'' strain RosettaBlue (DE3) and a short nucleotide structure from AP1 or AG promoter region. Final models of LFY-pAP1 and LFY-pAG were solved at 2.1 Å and 2.3 Å by X-ray diffraction and deposited as PDB entry <scene name='57/579703/2vy1/1'>2VY1</scene>/<scene name='57/579703/2vy2/1'>2VY2</scene>. |
=== Site specific DNA recognition is conducted by a HTH-like motif === | === Site specific DNA recognition is conducted by a HTH-like motif === | ||
The general structure of LEAFY DNA binding domain consists 2 <scene name='57/579703/Beta-strand/1'>β strands</scene> at the beginning followed by 7 <scene name='57/579703/Alpha-helices_color/2'> α helices</scene>. A <scene name='57/579703/Alpha-helices_color/3'>helix-turn-helix</scene> (HTH) motif can be found between α2 and α3 helices, which is recruited to the <scene name='57/579703/Major_groove/2'>major groove </scene>of the binding DNA. There are two amino acid at this motif, <scene name='57/579703/Major_groove_asn291/1'>Asn 291</scene> on α2 and <scene name='57/579703/Major_groove_asn291/2'>Lys 307</scene> on α3 directly mediate site specific recognition with <scene name='57/579703/Major_groove_detail/1'>two guanines</scene> at the DNA strand. These two recognition sites were further validated by electrophoresis mobility shift assay (EMSA): mutation at either Asn 291 or Lys 307 dramatically decrease binding affinity to pAP1. In the minor groove, site specific recognition is conducted by <scene name='57/579703/Arg_237/1'>Arg 237</scene>, which is at the beginning of this structure. ''Arabidopsis'' intermediate mutant ''lfy-4'' (P240L) and ''lfy-5'' (T244M) were located near this site and validate the function ''in planta''. The super position of specific recognition sites is summaries at right figure produced by PyMol. | The general structure of LEAFY DNA binding domain consists 2 <scene name='57/579703/Beta-strand/1'>β strands</scene> at the beginning followed by 7 <scene name='57/579703/Alpha-helices_color/2'> α helices</scene>. A <scene name='57/579703/Alpha-helices_color/3'>helix-turn-helix</scene> (HTH) motif can be found between α2 and α3 helices, which is recruited to the <scene name='57/579703/Major_groove/2'>major groove </scene>of the binding DNA. There are two amino acid at this motif, <scene name='57/579703/Major_groove_asn291/1'>Asn 291</scene> on α2 and <scene name='57/579703/Major_groove_asn291/2'>Lys 307</scene> on α3 directly mediate site specific recognition with <scene name='57/579703/Major_groove_detail/1'>two guanines</scene> at the DNA strand. These two recognition sites were further validated by electrophoresis mobility shift assay (EMSA): mutation at either Asn 291 or Lys 307 dramatically decrease binding affinity to pAP1. In the minor groove, site specific recognition is conducted by <scene name='57/579703/Arg_237/1'>Arg 237</scene>, which is at the beginning of this structure. ''Arabidopsis'' intermediate mutant ''lfy-4'' (P240L) and ''lfy-5'' (T244M) were located near this site and validate the function ''in planta''. The super position of specific recognition sites is summaries at right figure produced by PyMol. | ||
Revision as of 22:07, 18 May 2014
| This Sandbox is Reserved from 01/04/2014, through 30/06/2014 for use in the course "510042. Protein structure, function and folding" taught by Prof Adrian Goldman, Tommi Kajander, Taru Meri, Konstantin Kogan and Juho Kellosalo at the University of Helsinki. This reservation includes Sandbox Reserved 923 through Sandbox Reserved 947. |
To get started:
More help: Help:Editing |
Contents |
Evolution of DNA binding domain of LEAFY: from angiosperms to mosses
Introduction
FLORICAULA/LEAFY (FLO/LFY) genes encode a plant specific transcription factor family that controlling floral fate of reproductive phase. [1][2]. In the plant model system Arabidopsis thaliana , ‘’LFY’’ also acts upstream of floral homeotic genes to modulate organ identity. [3] LFY activates the organ identity genes by binding to promoter regions of floral organ identity genes. LFY can directly bind to the promoter to APELATA1 (AP1), while co-regulators UNUSUAL FLORAL ORGANS (UFO) [4] and WUSCHEL (WUS)[5] are required for increment of binding affinity to promoter regions of APELATA3 (AP3) and AGAMOUS (AG), respectively. The exact mechanism how LFY binds to these promoters has yet to be well elucidated until the first structure report about and [6] . Among land plants, FLO/LFY homologs share a highly conserved DNA binding region that a hypothesis claimed substitution in this domain might result in the functional divergence[7] . Recently, a new study provided new insights of structural basis of LEAFY evolution by changing DNA binding activity[8].
| |||||||||||
LEAFY Evolution
Reference
- ↑ 1.0 1.1 Detlef Weigel, John Alvarez, David R. Smyth, Martin F. Yanofsky, Elliot M. Meyerowitz, LEAFY controls floral meristem identity in Arabidopsis. Cell 69 :843-859, http://dx.doi.org/10.1016/0092-8674(92)90295-N.
- ↑ 2.0 2.1 Coen, E.S., Romero, J.M., Doyle, S., Elliot, R., Murphy, G. & Carpenter, R. (1990) floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 http://dx.doi.org/10.1016/0092-8674(90)90426-F
- ↑ Irish, V. F. (2010), The flowering of Arabidopsis flower development. The Plant Journal, 61: 1014–1028. http://dx.doi.org/10.1111/j.1365-313X.2009.04065.x
- ↑ Chae, E., Tan, Q.K., Hill, T.A. & Irish, V.F. 2008. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135:1235-45 http://dx.doi.org/10.1242/dev.015842
- ↑ HONG, R.L., HAMAGUCHI, L., BUSCH, M.A. and WEIGEL, D. (2003). Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15: 1296-1309. http://dx.doi.org/10.1105/tpc.009548
- ↑ Hames, C., Ptchelkine, D., Grimm, C., Thevenon, E., Moyroud, E., Gérard, F. Martiel, J.L., Benlloch, R., Parcy, F. & Müller, C.W. 2008. Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO Journal 27:2628-2637. http://dx.doi.org/10.1038/emboj.2008.184
- ↑ MAIZEL, A., BUSCH, M.A., TANAHASHI, T., PERKOVIC, J., KATAO, M., HASEBE, M. and WEIGEL, D. (2005). The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308: 260-263. http://dx.doi.org/10.1126/science.1108229
- ↑ Sayou, C., Monniaux, M., Nanao, M.H., Moyroud, E., Brockington, S.F., Thévenon, E., Chahtane, H., Warthmann, N., Melkonian, M., Zhang, Y., Wong, G., Weigel, D., Parcy, F. and Dumas, R. 2014. A Promiscuous Intermediate Underlies the Evolution of LEAFY DNA Binding Specificity Science 343: 645-648 http://dx.doi.org/10.1126/science.1248229
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedHames_et_al.