Ferredoxin
From Proteopedia
| Line 3: | Line 3: | ||
[[Ferredoxin]] (Fd) is found in chloroplasts which mediates electron transfer and contains an iron-sulfur cluster. It is involved in the photosynthesis process where its iron atoms accept or discharge electrons when they are being oxidized or reduced. The iron-sulfur cluster can contain 2Fe-2S and is termed plant-like or 3Fe-4S or 4Fe-4S clusters. '''Adrenodoxin''' (ADR) is a ferredoxin containing a 2Fe-2S group involved in electron transfer from NADPH+ to a cytochrome P-450 in the adrenal gland. '''Putidaredoxin''' (PUT) and '''terpredoxin''' (TER) are involved in the same reaction in bacteria and contain a 2Fe-2S group. | [[Ferredoxin]] (Fd) is found in chloroplasts which mediates electron transfer and contains an iron-sulfur cluster. It is involved in the photosynthesis process where its iron atoms accept or discharge electrons when they are being oxidized or reduced. The iron-sulfur cluster can contain 2Fe-2S and is termed plant-like or 3Fe-4S or 4Fe-4S clusters. '''Adrenodoxin''' (ADR) is a ferredoxin containing a 2Fe-2S group involved in electron transfer from NADPH+ to a cytochrome P-450 in the adrenal gland. '''Putidaredoxin''' (PUT) and '''terpredoxin''' (TER) are involved in the same reaction in bacteria and contain a 2Fe-2S group. | ||
| - | |||
| - | {{TOC limit|limit=2}} | ||
==D14C variant of ''Pyrococcus furiosus'' ferredoxin<ref>DOI 10.1007/s00775-011-0778-7</ref>== | ==D14C variant of ''Pyrococcus furiosus'' ferredoxin<ref>DOI 10.1007/s00775-011-0778-7</ref>== | ||
| Line 28: | Line 26: | ||
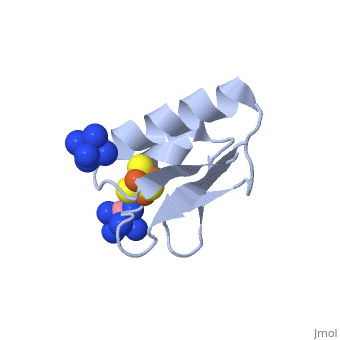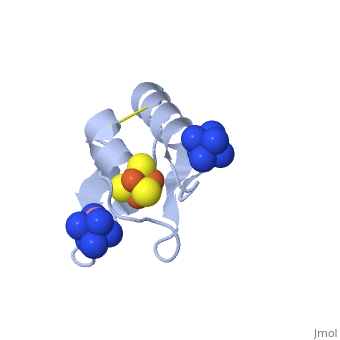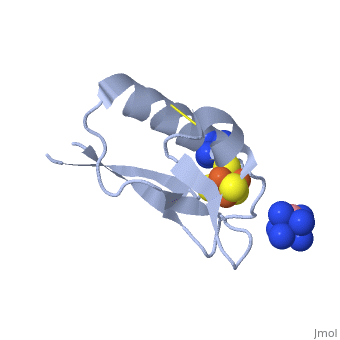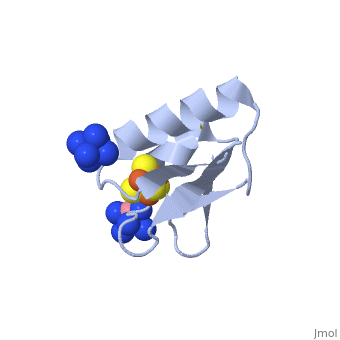
The crystal structure of the <scene name='Journal:JBIC:19/Cv/4'>monomeric form</scene> shows that the <scene name='Journal:JBIC:19/Cv/6'>silver (I) ion is part of the cluster</scene> (clearly seen on the electron density map), as predicted from previous spectroscopic and electrochemical studies. The heterometal is coordinated to the <scene name='Journal:JBIC:19/Cv/5'>three inorganic sulfides of the cluster</scene> and to the <scene name='Journal:JBIC:19/Cv/7'>thiolate group of residue 14</scene> (residues <scene name='Journal:JBIC:19/Cv/8'>Cys11, Cys17 and Cys56</scene> are coordinated with Fe ions of heterometal), <scene name='Journal:JBIC:19/Cv/9'>replacing the aboriginal Fe ion in the all-iron coordinated</scene> [Fe<sub>4</sub>S<sub>4</sub>] D14C variant ([[2z8q]], <span style="color:cyan;background-color:black;font-weight:bold;">heterometallic [AgFe<sub>3</sub>S<sub>4</sub>] protein is in cyan</span> and <span style="color:lime;background-color:black;font-weight:bold;">homometallic [Fe<sub>4</sub>S<sub>4</sub>] is in green</span>) and completing the incomplete cuboidal cluster present in the [Fe<sub>3</sub>S<sub>4</sub>] WT Pf Fd (PDB: [[1sj1]]) and its D14C (PDB: [[3pni]]) variant (for more details see also [http://proteopedia.org/w/Journal:JBIC:10 "Crystal structures of the all cysteinyl coordinated D14C variant of ''Pyrococcus furiosus'' ferredoxin: (4Fe-4S) <-> (3Fe-4S) cluster conversion"]). Structure alignment of backbone atoms from the heterometallic [AgFe<sub>3</sub>S<sub>4</sub>] protein and the homometallic [Fe<sub>4</sub>S<sub>4</sub>] D14C variant ([[2z8q]]) shows <scene name='Journal:JBIC:19/Cv/10'>very minor differences</scene>, i.e. the root mean square deviation (RMSD) is 0.4 – 0.7 Å, observed due to the alternate conformation of the main chain atoms, flexible loops and small changes at the N- and C-termini (<span style="color:cyan;background-color:black;font-weight:bold;">heterometallic [AgFe<sub>3</sub>S<sub>4</sub>] protein is in cyan</span> and <span style="color:lime;background-color:black;font-weight:bold;">homometallic [Fe<sub>4</sub>S<sub>4</sub>] is in green</span>). <scene name='Journal:JBIC:19/Cv/14'>More significant difference</scene> can be seen in the superimposed Fe–S clusters (atom colors corresponding to <span style="color:yellow;background-color:black;font-weight:bold;">yellow: S</span>, <span style="color:orange;background-color:black;font-weight:bold;">orange: Fe</span>, <span style="color:gray;background-color:black;font-weight:bold;">gray: Ag</span>) from these two variants, which is due to the presence of the second row transition metal ion (Ag) coordinated to the four S-ligands, i.e. the presence of Ag results in a distorted geometry of the cluster compared to the all-iron arrangement, <scene name='Journal:JBIC:19/Cv/12'>because to the longer</scene> Ag – S bond lengths compared to Fe – S bonds. However, the S – Ag – S <scene name='Journal:JBIC:19/Cv/13'>bond angles</scene> are still close to the expected 90° for a primitive cubic system. | The crystal structure of the <scene name='Journal:JBIC:19/Cv/4'>monomeric form</scene> shows that the <scene name='Journal:JBIC:19/Cv/6'>silver (I) ion is part of the cluster</scene> (clearly seen on the electron density map), as predicted from previous spectroscopic and electrochemical studies. The heterometal is coordinated to the <scene name='Journal:JBIC:19/Cv/5'>three inorganic sulfides of the cluster</scene> and to the <scene name='Journal:JBIC:19/Cv/7'>thiolate group of residue 14</scene> (residues <scene name='Journal:JBIC:19/Cv/8'>Cys11, Cys17 and Cys56</scene> are coordinated with Fe ions of heterometal), <scene name='Journal:JBIC:19/Cv/9'>replacing the aboriginal Fe ion in the all-iron coordinated</scene> [Fe<sub>4</sub>S<sub>4</sub>] D14C variant ([[2z8q]], <span style="color:cyan;background-color:black;font-weight:bold;">heterometallic [AgFe<sub>3</sub>S<sub>4</sub>] protein is in cyan</span> and <span style="color:lime;background-color:black;font-weight:bold;">homometallic [Fe<sub>4</sub>S<sub>4</sub>] is in green</span>) and completing the incomplete cuboidal cluster present in the [Fe<sub>3</sub>S<sub>4</sub>] WT Pf Fd (PDB: [[1sj1]]) and its D14C (PDB: [[3pni]]) variant (for more details see also [http://proteopedia.org/w/Journal:JBIC:10 "Crystal structures of the all cysteinyl coordinated D14C variant of ''Pyrococcus furiosus'' ferredoxin: (4Fe-4S) <-> (3Fe-4S) cluster conversion"]). Structure alignment of backbone atoms from the heterometallic [AgFe<sub>3</sub>S<sub>4</sub>] protein and the homometallic [Fe<sub>4</sub>S<sub>4</sub>] D14C variant ([[2z8q]]) shows <scene name='Journal:JBIC:19/Cv/10'>very minor differences</scene>, i.e. the root mean square deviation (RMSD) is 0.4 – 0.7 Å, observed due to the alternate conformation of the main chain atoms, flexible loops and small changes at the N- and C-termini (<span style="color:cyan;background-color:black;font-weight:bold;">heterometallic [AgFe<sub>3</sub>S<sub>4</sub>] protein is in cyan</span> and <span style="color:lime;background-color:black;font-weight:bold;">homometallic [Fe<sub>4</sub>S<sub>4</sub>] is in green</span>). <scene name='Journal:JBIC:19/Cv/14'>More significant difference</scene> can be seen in the superimposed Fe–S clusters (atom colors corresponding to <span style="color:yellow;background-color:black;font-weight:bold;">yellow: S</span>, <span style="color:orange;background-color:black;font-weight:bold;">orange: Fe</span>, <span style="color:gray;background-color:black;font-weight:bold;">gray: Ag</span>) from these two variants, which is due to the presence of the second row transition metal ion (Ag) coordinated to the four S-ligands, i.e. the presence of Ag results in a distorted geometry of the cluster compared to the all-iron arrangement, <scene name='Journal:JBIC:19/Cv/12'>because to the longer</scene> Ag – S bond lengths compared to Fe – S bonds. However, the S – Ag – S <scene name='Journal:JBIC:19/Cv/13'>bond angles</scene> are still close to the expected 90° for a primitive cubic system. | ||
</StructureSection> | </StructureSection> | ||
| - | + | ||
===3D structures of ferredoxin=== | ===3D structures of ferredoxin=== | ||
Revision as of 08:47, 14 August 2014
| |||||||||||
Contents |
3D structures of ferredoxin
Updated on 14-August-2014
2Fe-2S containing ferredoxins
2kaj, 1dox, 1doy – SyFd +Ga – Synechocystis – NMR
3hui – Fd – Rhodopseudomonas palustris
1off – SyFd
3gce – Fd – Nocardioides aromaticivorans
3lxf – Fd – Novosphingobium aromaticivorans
2e4p, 2e4q - Fd – Pseudomonas sp.
2q3w, 1vm9 - PmFd (mutant) – Pseudomonas mendocina
2i7f, 1uwm - RcFd – Rhodobacter capsulatus
1e9m – RcFd - NMR
1rfk, 3p63 - Fd – Cyanobacterium masticogladus laminosus
1vck - Fd – Pseudomonas resinovorans
1wri, 1frr - Fd – Equisetum arvense
1sjg - PmFd– NMR
1iue - Fd– Plasmodium falciparum
1m2a – AeFd – Aquifex aeolicus
1m2b, 1m2d, 1f37, 1f5b, 1f5c – AeFd (mutant)
1l5p – Fd – Trichomonas vaginalis
1i7h - Fd – Escherichia coli
1czp, 1qt9, 1frd, 1fxa - aFd– anabaena
1j7a, 1j7b, 1j7c , 1qoa, 1qob, 1qof, 1qog- aFd (mutant)
1e0z – Fd – Halobacterium salinarium
1e10 - Fd – Halobacterium halobium
1pfd – Fd – Petroselinum crispum – NMR
1a70 - Fd (mutant) – Spinacia oleracea
1awd - Fd – Chlorella fusca
2cjn, 2cjo, 1roe – SyFd – NMR
1rof – SyFd – Synechococcus elongates
1doi - Fd – Haloarcula marismortui
4fxc – Fd – Spirulina platensis
1fxi, 3av8 – Fd – Aphanothece sacrum
3dqy, 2qpz, 3ah7 - Fd – Pseudomonas putida
3ab5, 3wcq – Fd – Cyanidioschyzon merolae
3b2f – mFd – maize
3w5u, 3w5v – mFd + ferredoxin-NADP reductase
3b2g – Fd – Leptolyngbya boryana
4Fe-4S containing ferredoxins
3eun – AvFd – Allochromatium vinosum
3exy - AvFd (mutant)
2vkr - Fd+Zn – Acidianus ambivalens
2z8q - PfFd (mutant) – Pyrococcus furiosus
1siz - PfFd
3pni - PfFd (mutant)
2fgo - Fd– Pseudomonas aeruginosa
1iqz, 1ir0 - BtFd – Bacillus thermoproteolyticus
1rgv - Fd – Thauera aromatica
1dax, 1dfd – DaFd – Desulfovibrio africanus – NMR
1fxr - DaFd
1vjw – Fd – Thermotoga maritima
1dwl – Fd – Desulfomicrobium norvegicum
3Fe-4S containing ferredoxins
2v2k – Fd – Mycobacterium smegmatis
1wtf - BtFd (mutant)
1sj1 - PfFd
1fxd - DgFd – Desulfovibrio gigas
1f2g – DgFd – NMR
1xer - Fd – Sulfolobus tokodaii
4Fe-4S+3Fe-4S containing ferredoxins
1gao, 6fdr, 7fd1, 7fdr, 1axq, 6fd1, 1frh, 1fri,1frj, 1frk, 1frl, 1frm, 1fda, 1fdb, 1fdd, 5fd1, 1fer – AvFd – Azotobacter vinelandii
1pc4, 1pc5, 1g6b, 1g3o, 1ff2, 1b0v, 1d3w, 1b0t, 1a6l, 1ftc, 1frx, 2fd2, 1fd2 - AvFd (mutant)
1h98 – Fd – Thermus thermophilus
1a8p, 1bd6, 1bc6 - BsFd – Bacillus schlegelii – NMR
4Fe-4S+4Fe-4S containing ferredoxins
1dur – Fd – Peptoniphilus asaccarolyticus
1bwe, 1bqx - BsFd (mutant) – NMR
2fdn, 1fca , 1fdn- Fd – Clostridium acidi-urici
1blu - Fd – Chromatium vinosum
1clf – Fd – Clostridium pasteurianum – NMR
Adrenoredoxin
2jqr – ADR Fd domain (mutant)+cytochrome c (mutant) – yeast – NMR
2bt6 – cADR1 modified – cow
1l6u, 1l6v – cADR1 – NMR
1e6e – cADR (mutant)+ADR reductase
1cje, 1ayf - cADR
3na0, 3n9y, 3n9z, 3na1 - hADR +cholesterol side-chain cleavage enzyme – human
3p1m - hADR
Putidaredoxin
1yji, 1yjj, 1pdx – PpPUT – NMR
3lb8 – PpPUT (mutant)+PUT reductase
1xln, 1xlo, 1xlp, 1xlq, 1r7s, 1oqq, 1oqr - PpPUT (mutant)
1gpx, 1put- PpPUT (mutant) - NMR
Terpredoxin
1b9r – TER – Pseudomonas - NMR
References
- ↑ Lovgreen MN, Martic M, Windahl MS, Christensen HE, Harris P. Crystal structures of the all-cysteinyl-coordinated D14C variant of Pyrococcus furiosus ferredoxin: [4Fe-4S] <--> [3Fe-4S] cluster conversion. J Biol Inorg Chem. 2011 Apr 12. PMID:21484348 doi:10.1007/s00775-011-0778-7
- ↑ Iwasaki T, Kappl R, Bracic G, Shimizu N, Ohmori D, Kumasaka T. ISC-like [2Fe-2S] ferredoxin (FdxB) dimer from Pseudomonas putida JCM 20004: structural and electron-nuclear double resonance characterization. J Biol Inorg Chem. 2011 Jun 7. PMID:21647778 doi:10.1007/s00775-011-0793-8
- ↑ Martic M, Jakab-Simon IN, Haahr LT, Hagen WR, Christensen HE. Heterometallic [AgFe(3)S (4)] ferredoxin variants: synthesis, characterization, and the first crystal structure of an engineered heterometallic iron-sulfur protein. J Biol Inorg Chem. 2013 Feb;18(2):261-76. doi: 10.1007/s00775-012-0971-3. Epub, 2013 Jan 8. PMID:23296387 doi:10.1007/s00775-012-0971-3
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky, Eran Hodis, Wayne Decatur, David Canner
![pH dependent equilibrium of D14C [3Fe-4S] P. furiosus ferredoxin between protonated and deprotonated monomers and formation of a disulfide bonded dimer from deprotonated monomers. Fd is short for ferredoxin.](/wiki/images/thumb/0/0b/Schem1.png/300px-Schem1.png)
![Skewed orientations of the gmax component (red) with respect to the molecular frame of the [2Fe–2S] cluster of FdxB.](/wiki/images/2/29/FdxBFig8.jpg)
